Add TCs to historical data
Contents
12. Add TCs to historical data#
Results of simulated historical TCs (SHyTCWaves) are added to historical data (surges and waves)
inputs required:
Historical DWTs
Historical storm surge
Historical super-point spectra
Historical bulk parameters
Track and pressures of historical TCs simulated with SHyTCWaves (for storm surge)
Super-point of historical TCs simulated with SHyTCWaves (for waves)
in this notebook:
Add surge from TCs whenever the simulated TC enters the 7.5 degrees radio.
Add waves spectrum from TCs whenever the simulated TC enters the 7.5 degrees radio.
The historical generation of surge and wave conditions on a TC climate (DWT37-42) is made by checking if a TC that affects the site on a 14º radio also affects the site on a 7.5º radio; if that is the case, the storm surge (inverse barometer) and the super-point spectrum from the TC will be incorporated, replacing the surge for the day when the TC is closest to site and the spectrum for the complete duration of the TC.
#!/usr/bin/env python
# -*- coding: utf-8 -*-
# common
import os
import os.path as op
import glob
import sys
# pip
import numpy as np
import xarray as xr
import pandas as pd
from datetime import datetime
import matplotlib.pyplot as plt
from matplotlib import gridspec
import string
import pickle5 as pk
import wavespectra
# DEV: override installed teslakit
sys.path.insert(0, op.join(os.path.abspath(''), '..', '..', '..'))
# teslakit
from bluemath.teslakit.io.aux_nc import StoreBugXdset
from bluemath.teslakit.waves_emulator import add_TC_to_historical
Warning: ecCodes 2.21.0 or higher is recommended. You are running version 2.20.0
/Users/albacid/Software/Bitbucket_repos/teslakit.2.0/venv/lib/python3.7/site-packages/wavespectra/specarray.py:29: UserWarning: Cannot import sympy, install "extra" dependencies for full functionality
'Cannot import sympy, install "extra" dependencies for full functionality'
12.1. Files and paths#
# project path
p_data = '/Users/albacid/Projects/TeslaKit.2.0_projects/SAMOA'
site = 'samoa'
# waves from historical and synthetic TCs in r2
p_TCs ='/Volumes/WD/SHyTCWaves_samoa/shy_tesla_samoa_historic'
# input files
dwt_his_file = op.join(p_data,'ESTELA', 'pred_slp_grd', 'kma.nc')
IB_file = op.join(p_data, 'TIDE','IB_noTCs_daily_his.nc')
superpoint_file = op.join(p_data,'WAVES','Super_point_superposition_15.nc')
bulk_his_file = op.join(p_data,'WAVES','bulk_params_wcut_1e-11.nc')
tcs_hist_r1_params_file = op.join(p_data,'TCs','TCs_hist_r1_params_' + site + '_raw.nc') # historical TCs parameters r1
tcs_hist_r1_params_file_sel = op.join(p_data,'TCs','TCs_hist_r1_params_' + site + '.nc')
tcs_r2_file = op.join(p_data, 'resources','TCs_ShyTCwaves','indices_histnak_' + site + '_r8.nc') # TCs in r2 (shyTCwaves)
# output file
IB_TCs_file = op.join(p_data, 'TIDE', 'IB_TCs_daily_his.nc')
spec_TCs_file = op.join(p_data, 'WAVES', 'Super_point_superposition_15_TCs.nc')
bulk_TCs_file = op.join(p_data, 'WAVES', 'bulk_params_wcut_1e-11_TCs.nc')
12.2. Parameters#
# Load historical surge
IB_noTCs = xr.open_dataset(IB_file)
# Load historical spectrum
spec_his = xr.open_dataset(superpoint_file)
_, index = np.unique(spec_his['time'], return_index=True)
spec_his = spec_his.isel(time=index)
# Load his DWTs
DWTs_his = xr.open_dataset(dwt_his_file)
DWTs_his = xr.Dataset({'bmus':(('time'), DWTs_his.sorted_bmus_storms.values)},
coords = {'time':(('time'), DWTs_his.time.values)})
# Load historical TCs from r1
TCs_his = xr.open_dataset(tcs_hist_r1_params_file)
TCs_his['id'] = (('storm'),np.arange(0, len(TCs_his.storm)))
TCs_his = TCs_his[['category','id','dmin_date']]
# filter TCs happening in consecutive days
TCs_his_filtered = xr.open_dataset(tcs_hist_r1_params_file_sel)
TCs_his = TCs_his.sel(storm=TCs_his_filtered.storm.values[:])
# add needed variables
TCs_his = TCs_his.assign_coords(storm = np.arange(0, len(TCs_his.storm)))
TCs_his['type'] = (('storm'), np.zeros((len(TCs_his.storm))))
TCs_his['type'][:] = 1
TCs_his['dmin_date_d'] = (('storm'), TCs_his.dmin_date.values.astype('datetime64[D]'))
# Load TCs from r2 (simulated TCs, shytcwaves)
tcs_r2 = xr.open_dataset(tcs_r2_file)
tcs_r2 = tcs_r2[['id_hist']]
12.3. Add TCs storm surge to historical surge#
# define output dataset
IB_TCs = IB_noTCs.copy(deep=True)
# variables to save when DWT from TCs
IB_TCs['TC_cat'] = (('time'), np.full((len(IB_TCs.time)), np.nan))
IB_TCs['TC_id'] = (('time'), np.full((len(IB_TCs.time)), np.nan))
# data for each day
for ind_t, (t, dwt) in enumerate(zip(DWTs_his.time.values, DWTs_his.bmus.values)):
if dwt>35:
print(t)
#------------------------------
# Select TC
TCs_his_t = TCs_his.where(TCs_his.dmin_date_d==t, drop=True)
# Check if r1 TC (TCs for obtaining DWTs) enters in r2 (simulated TCs)
tcs_r2_r1 = tcs_r2.where(tcs_r2.id_hist==TCs_his_t.id.values, drop=True)
tc_in_r2 = len(tcs_r2_r1.hist)
if tc_in_r2!=0 :
# Load historical TC track
file = glob.glob(op.join(p_TCs,'track_samoa_cat*_id' + str(int(TCs_his_t.id.values)) + '_historic.pkl'))
trackTC = pk.load(open(file[0],'rb'))
# Add IB from when TC passes closest to site
min_dist = np.argmin(trackTC.dist2land.values)
press_min_dist = trackTC.p0[min_dist]
IB_TC_mindist = press_min_dist - 1013
IB_TCs['level_IB'][ind_t] = (-1*IB_TC_mindist)/100.0
# Save selected TC indicators
IB_TCs['TC_cat'][ind_t] = TCs_his_t.category.values[0]
IB_TCs['TC_id'][ind_t] = TCs_his_t.id.values[0]
else:
print('TC not in r2')
# save
#IB_TCs.to_netcdf(IB_TCs_file)
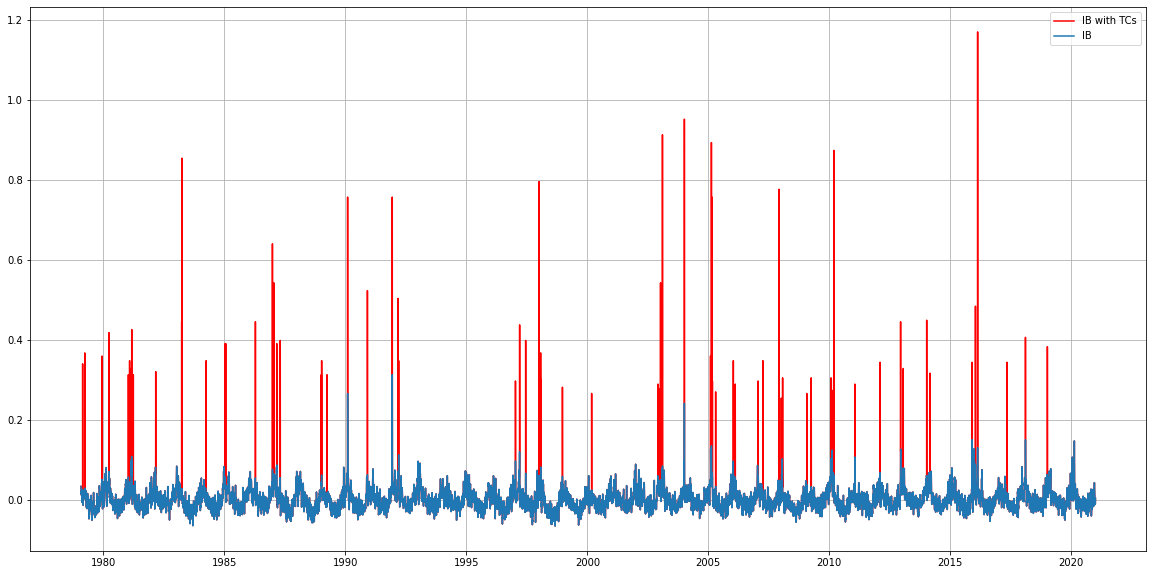
# Plot Inverse barometer of regular climate and TCs
surge = xr.open_dataset(IB_file)
surge_TCs = xr.open_dataset(IB_TCs_file)
plt.figure(figsize=(20,10))
plt.plot(surge_TCs.time, surge_TCs['level_IB'], 'r',label = 'IB with TCs')
plt.plot(surge.time, surge['level_IB'], label = 'IB')
plt.legend()
plt.grid()
#plt.xlim(datetime(2005,1,1), datetime(2006,1,1))
12.4. Add TCs spectrum to historical spectrum#
DWTs_n = DWTs_his.sel(time=slice(spec_his.time.values[0],spec_his.time.values[-1]))
# variables to save when DWT from TCs
spec_his['TC_cat'] = (('time'), np.full((len(spec_his.time)), np.nan))
spec_his['TC_id'] = (('time'), np.full((len(spec_his.time)), np.nan))
# data for each day
for t, dwt in zip(DWTs_n.time.values, DWTs_n.bmus.values):
if dwt>35:
print(t)
#------------------------------
# Select TC
TCs_his_t = TCs_his.where(TCs_his.dmin_date_d==t, drop=True)
# Check if r1 TC (TCs for obtaining DWTs) enters in r2 (simulated TCs)
tcs_r2_r1 = tcs_r2.where(tcs_r2.id_hist==TCs_his_t.id.values, drop=True)
tc_in_r2 = len(tcs_r2_r1.hist)
if tc_in_r2!=0 :
file_TC = glob.glob(op.join(p_TCs, 'superpoint_samoa_cat*_id' + str(int(TCs_his_t.id.values)) + '_historic.nc'))
# Add TC spectrum to regular climate spectrum
spec_TC = xr.open_dataset(file_TC[0])
spec_his = add_TC_to_historical(spec_TC, spec_his, TCs_his_t)
else:
print('TC not in r2')
#------------------------------
# compute bulk params
bulk_TCs = spec_his.spec.stats(['hs','tp','tm02','dpm','dspr'])
#------------------------------
# save bulk and spectrum
bulk_TCs['TC_cat'] = spec_his['TC_cat']
bulk_TCs['TC_id'] = spec_his['TC_id']
bulk_TCs.to_netcdf(bulk_TCs_file)
spec_his.to_netcdf(spec_TCs_file)
12.4.1. Add offset to Hs#
# add offset to TC bulk data
bulk = xr.open_dataset(bulk_his_file)
bulk_TCs = xr.open_dataset(bulk_TCs_file)
_, index = np.unique(bulk['time'], return_index=True)
bulk = bulk.isel(time=index)
# correct bulk
offset = np.mean(bulk.hs.values)
Hs = xr.where(~np.isnan(bulk_TCs.TC_cat.values), np.sqrt(offset**2 + bulk_TCs.hs.values**2), bulk_TCs.hs)
bulk_TCs['hs'] = (('time'), Hs)
# save
os.remove(bulk_TCs_file)
bulk_TCs.to_netcdf(bulk_TCs_file)
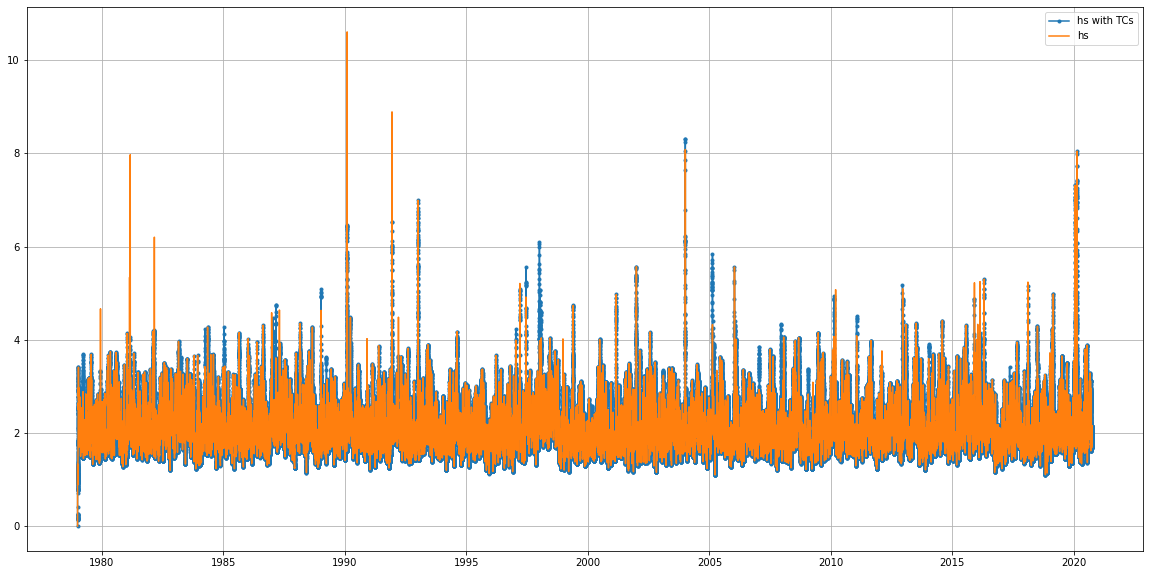
var = 'hs'
# plot time series
plt.figure(figsize=(20,10))
plt.plot(bulk_TCs.time, bulk_TCs[var], '.-',label = var + ' with TCs')
plt.plot(bulk.time, bulk[var], label = var)
plt.legend()
plt.grid()
#plt.xlim(datetime(1990,1,1), datetime(1990,3,1))
12.4.2. Add offset to spectrum#
# Load spectrum
spec = xr.open_dataset(superpoint_file)
spec_TCs = xr.open_dataset(spec_TCs_file)
# correct spectrum
offset = spec.efth.mean(dim='time')
ind = np.where(~np.isnan(spec_TCs.TC_cat.values))[0]
spec_TCs['efth'][ind,:,:] = offset.values + spec_TCs.efth.values[ind,:,:]
# save
os.remove(spec_TCs_file)
spec_TCs.to_netcdf(spec_TCs_file)

