Conditioned probabilities between Predictor and Predictand
Contents
Conditioned probabilities between Predictor and Predictand#
# common
import warnings
warnings.filterwarnings('ignore')
import os
import os.path as op
import sys
# pip
import numpy as np
import xarray as xr
# DEV: bluemath
sys.path.insert(0, op.join(op.abspath(''), '..', '..', '..', '..', '..'))
# bluemath modules
from bluemath.kma import calculate_conditioned_probabilities
from bluemath.binwaves.plotting.classification import Plot_spectra_hs_pcs, Plot_kmeans_clusters_hs
from bluemath.plotting.slp import Plot_slp_kmeans
from bluemath.plotting.teslakit import Plot_DWTs_Probs, Plot_Probs_WT_WT
Database and site parameters#
# database
p_data = r'/media/administrador/HD2/SamoaTonga/data'
site = 'Samoa'
p_site = op.join(p_data, site)
# deliverable folder
p_deliv = op.join(p_site, 'd04_seasonal_forecast_swells')
p_kma = op.join(p_deliv, 'kma')
# superpoint classification
p_kma_spec_classification = op.join(p_kma, 'spec_KMA_classification.nc')
# estela classification
p_kma_slp_classification = op.join(p_kma, 'slp_KMA_classification.nc')
# output file: conditioned probabilities
p_probs_cond = op.join(p_deliv, 'conditioned_prob_SLP_SPEC.nc')
Predictand summary#
Note
This is a summary of the predictand classification developed before
spec_kma = xr.open_dataset(p_kma_spec_classification)
num_clusters_spec = len(np.unique(spec_kma.bmus))
f1, mean_h_clust, hs_m = Plot_kmeans_clusters_hs(spec_kma, vmax=1.5);
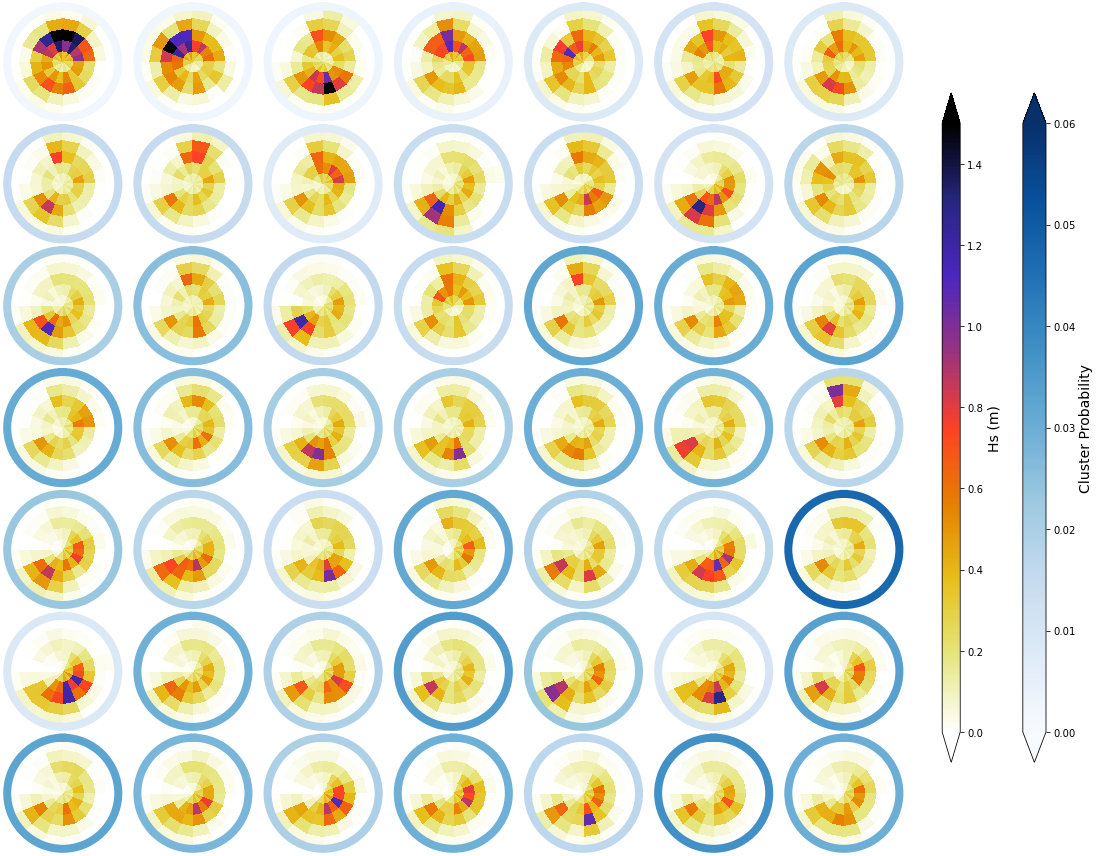
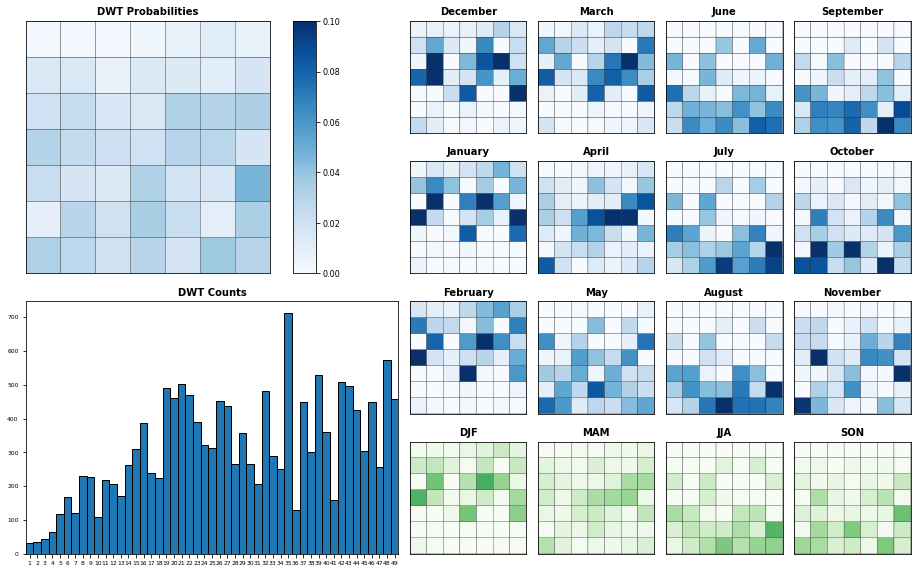
Plot_DWTs_Probs(spec_kma.bmus+1, spec_kma.time.values, num_clusters_spec, vmax=0.08);
Predictor summary#
Note
This is a summary of the predictor classification developed before
slp_kma = xr.open_dataset(p_kma_slp_classification)
num_clusters_slp = len(np.unique(slp_kma.bmus))
f2 = Plot_slp_kmeans(slp_kma, extent=(50, 290, -67, 65), figsize=[23,14]);

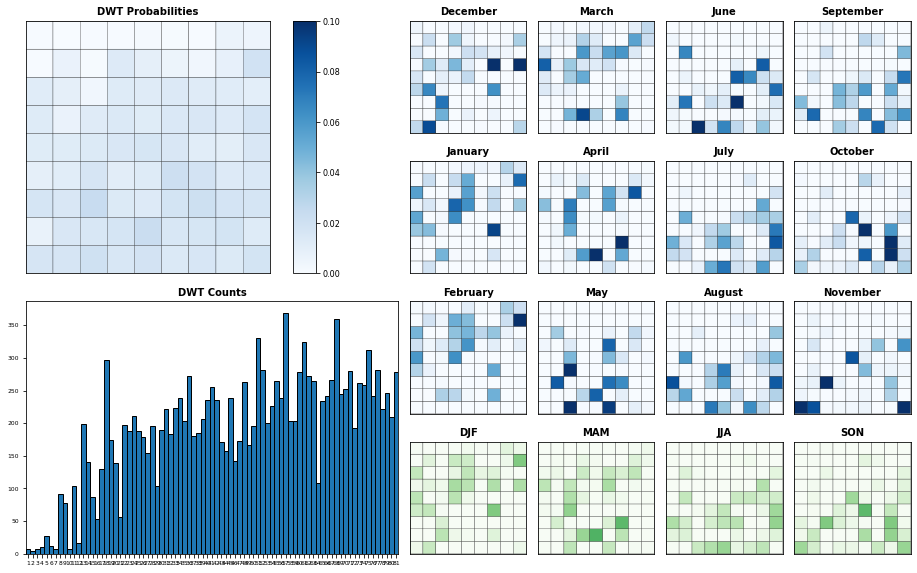
Plot_DWTs_Probs(slp_kma.bmus+1, slp_kma.time.values, num_clusters_slp, vmax=0.1);
Conditioned probabilities#
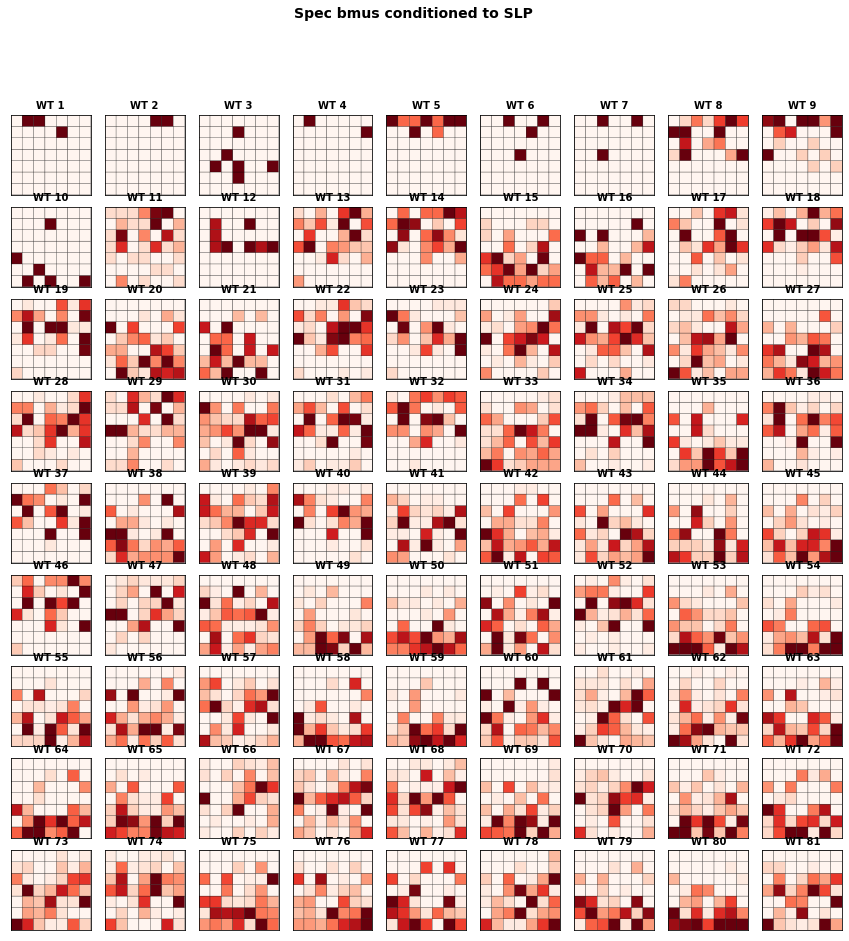
The figure below represents the conditioned probability of the 49 predictand clusters, conditioned to the 81 predictor clusters.
We observe large variability on the probability distributions of spectral clusters associated to each SLP cluster.
c, i_sp, i_slp = np.intersect1d(spec_kma.time, slp_kma.time, return_indices=True)
slp_kma = slp_kma.isel(time = i_slp)
spec_kma = spec_kma.isel(time = i_sp)
f3 = Plot_Probs_WT_WT(
slp_kma.bmus, spec_kma.bmus,
num_clusters_slp, num_clusters_spec,
wt_colors = False,
ttl = 'Spec bmus conditioned to SLP',
figsize = [15,15],
vmax = 0.07,
);
# calculate conditioned probabilities
conditioned_prob = calculate_conditioned_probabilities(slp_kma, num_clusters_slp, spec_kma, num_clusters_spec)
conditioned_prob.to_netcdf(p_probs_cond)

