HyFlood plots for Samoa
Contents
HyFlood plots for Samoa#
Plot the flood map of a given Hypercube-sampled case, for a transect in a mesh.
Needed data:
Flood cases in pickle format.
These are the outputs from Lisflood-FP runs, which contain a set of XYZ points, where Z is the maximum elevation obtained in that coordinate for the case.Digital Elevation Model (DEM) in NetCDF format.
The name coding is as follows: “XL_N.nc” where X is the initial letter of the island and N is the mesh number. Therefore, “UL_04.nc” would be the fourth mesh of Upolu island.Synthetic events in NetCDF format.
It contains a xarray.Dataset of 50 cases defined by their volume (V), their maximum level (Z*), their IG wave amplitude (Asig) and their duration (D). This cases are generated following the Latin Hypercube Sampling technique, which generates a random multidimensional set of parameters by arbitrary discretizing each dimension in N equally probable intervals.
Shoreline in pickle format.
It contains an array of float64 with X, Y and transect ID of every shoreline point of the selected site.
import os
import os.path as op
import sys
import numpy as np
import pandas as pd
import xarray as xr
import pickle
from natsort import natsorted
import matplotlib.pyplot as plt
import cmocean as cmo
import copy
from HyFlood_plots.plots import *
from ipywidgets import FloatProgress
from IPython.display import display
from time import sleep
Select location#
Selection of the desired site. There are two possible islands: Savaii and Upolu.
Each island is divided in meshes that have variable size. Savaii has 9 meshes, Upolu have 11.
Each island also has a different number of transects. Savaii has 322 transects and Upolu has 452.
The Highest Astronomical Tide in Samoa is 0.738m above Mean Sea Level.
Parameters#
Site : Desired island
Mesh : Mesh ID
Transect : Transect ID
#-----------------------------------------------------------
site = 'upolu' # ['upolu' or 'savaii']
mesh = 3 # Check the number of meshes of your island (above)
transect = 50 # Check the transects for your island (above)
#-----------------------------------------------------------
# Coordinate system and name code
epsg = 32702
if site == 'savaii':
code = 'S'
else:
code = 'U'
hat = 0.738 # Highest Astronomical Tide
# INPUT PATHS
pdat = op.abspath(op.join(os.getcwd(), '..'))
pcas = op.join(pdat,'compressed_outputs_{0}L_{1:02d}'.format(code,mesh))
pdem = op.join(pdat,'data/sites/{0}/meshes'.format(site))
phyd = op.join(pdat,'data/sites/{0}/forcings'.format(site))
psho = op.join(pdat,'data/sites/{0}/shoreline'.format(site))
# OUTPUT PATHS
pfig = op.join(os.getcwd(),'figures/')
if not op.isdir(pfig): os.makedirs(pfig)
# LOAD DATA
print('Island: {0} \t Mesh: {1}L_{2:02d} \t Transect: {3} \t \n'.format(site,code,mesh,transect))
# Floods
wds = []
for root, _, file in os.walk(pcas):
for name in file:
if name.startswith('max_t{0:03d}'.format(transect)):
wds.append(name)
wds = natsorted(wds)
print('Cases in the transect: {0} \n'.format(len(wds)))
print(wds)
# DEM
dem = xr.open_dataset(op.join(pdem,'{0}L_{1:02d}.nc'.format(code,mesh)))
# Forcings
hyd = xr.open_dataset(op.join(phyd,'cases50_IG.nc'))
# Shoreline
sh = pd.read_pickle(op.join(psho, 'shore_{0}_transects.pkl'.format(site)))
Island: upolu Mesh: UL_03 Transect: 50
Cases in the transect: 50
['max_t050_case00.pkl', 'max_t050_case01.pkl', 'max_t050_case02.pkl', 'max_t050_case03.pkl', 'max_t050_case04.pkl', 'max_t050_case05.pkl', 'max_t050_case06.pkl', 'max_t050_case07.pkl', 'max_t050_case08.pkl', 'max_t050_case09.pkl', 'max_t050_case10.pkl', 'max_t050_case11.pkl', 'max_t050_case12.pkl', 'max_t050_case13.pkl', 'max_t050_case14.pkl', 'max_t050_case15.pkl', 'max_t050_case16.pkl', 'max_t050_case17.pkl', 'max_t050_case18.pkl', 'max_t050_case19.pkl', 'max_t050_case20.pkl', 'max_t050_case21.pkl', 'max_t050_case22.pkl', 'max_t050_case23.pkl', 'max_t050_case24.pkl', 'max_t050_case25.pkl', 'max_t050_case26.pkl', 'max_t050_case27.pkl', 'max_t050_case28.pkl', 'max_t050_case29.pkl', 'max_t050_case30.pkl', 'max_t050_case31.pkl', 'max_t050_case32.pkl', 'max_t050_case33.pkl', 'max_t050_case34.pkl', 'max_t050_case35.pkl', 'max_t050_case36.pkl', 'max_t050_case37.pkl', 'max_t050_case38.pkl', 'max_t050_case39.pkl', 'max_t050_case40.pkl', 'max_t050_case41.pkl', 'max_t050_case42.pkl', 'max_t050_case43.pkl', 'max_t050_case44.pkl', 'max_t050_case45.pkl', 'max_t050_case46.pkl', 'max_t050_case47.pkl', 'max_t050_case48.pkl', 'max_t050_case49.pkl']
# PREPROCESS
# Mesh limits
#xmin, xmax = np.amin(dem.x.values), np.amax(dem.x.values)
#ymin, ymax = np.amin(dem.y.values), np.amax(dem.y.values)
# Apia limits
#xmin, xmax = [413500,417000]
#ymin, ymax = [8470000,8473000]
# South Zone limits
xmin, xmax = [405800,407000]
ymin, ymax = [8474000,8475000]
# Cut shore
sx = np.where((sh[:,0] >= xmin) & (sh[:,0] <= xmax))[0]
sht = sh[sx]
sy = np.where((sht[:,1] >= ymin) & (sht[:,1] <= ymax))[0]
sht = sht[sy]
# Get transects inside the domain
transects = np.unique(sht[:,2])
print('Check if your transect is one of these: \n')
print(transects)
# Smooth DEM (if not done, plot can be computationally high-demanding)
dems = dem.isel(x=slice(None, None, 2), y=slice(None, None, 2))
Check if your transect is one of these:
[49. 50. 51. 52.]
HyFlood methodology#
In each transect of the selected mesh (except for the last and first 3, in order to avoid numerical instabilities), the 50 synthetic cases are simulated.
Boundary conditions for Lisflood-FP are the synthetic representation of a water-level-induced flood event. To achieve this, water level is considered as the linear combination of the highest astronomical tide, the maximum water elevation in the event and the amplitude of the associated IG wave:

Therefore, the synthetic event follows a equation of the form:
where \({\eta}*\) represents the free surface over the HAT. Thus, for each hour, free surface can be defined as a linear combination of \({\eta}*\) and \({{\eta_{IG}}}\). Events are then defined as the peaks over over a thershold (in this case the HAT):

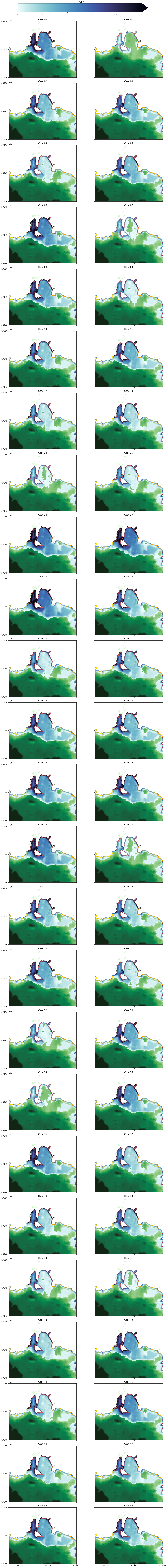
Plotting flood maps#
Here all flood extents are plotted for the indicated transect. The variable here shown is water depth in meters.
Notice that vmin and vmax values are defined inside the function “plot_one_trs”, along with other visual parameters such as colormaps. “cols” variable define the number of columns the figure will have.
# Domain limits
limits = [xmin,xmax,ymin,ymax]
fig, axs = plt.subplots(nrows=25, ncols=2, sharex=True, sharey=True, figsize = (17,140))
print('Plotting...')
fp = FloatProgress(min=0, max=49)
display(fp)
for idx, i in enumerate(wds[:]):
mxe = pd.read_pickle(op.join(pcas,i))
# Preprocess for plotting
mxe = mxe.reset_index(level=0)
mxe = mxe.reset_index(level=0)
colorwd, axp = plot_one_trs(axs,dems,mxe,sht,limits,idx)
sleep(0.05)
fp.value = idx
print('Done! Figure is almost ready')
blockPrint()
fig.subplots_adjust(wspace=0.05,hspace=0.1,top=0.99)
cbar_ax = fig.add_axes([0.2, 0.995, 0.6, 0.005])
cbar = plt.colorbar(colorwd, cax=cbar_ax, extend='max', orientation='horizontal')
cbar.ax.xaxis.set_label_position('top')
cbar.ax.set_xlabel('WD [m]')
enablePrint()
Plotting...
Done! Figure is almost ready