BinWaves & SWAN
Contents
BinWaves & SWAN#
import warnings
warnings.filterwarnings('ignore')
# common
import os
import os.path as op
import sys
import pickle as pk
import shutil
# pip
import numpy as np
import pandas as pd
import xarray as xr
# DEV: bluemath
sys.path.insert(0, op.join(op.abspath(''), '..', '..', '..', '..'))
# bluemath modules
from bluemath.common import store_xarray_max_compression
from bluemath.binwaves.plotting.grid import Plot_grid_cells, Plot_grid_cases
from bluemath.binwaves.plotting.swan_case import Plot_swan_case
from bluemath.wswan.wrap import SwanProject, SwanMesh, SwanWrap_STAT
from bluemath.wswan.plots.stationary import scatter_maps
Methodology#
BinWaves
A new methodology for efficiently propagating multivariate wave systems to nearshore locations is propossed.
The methodology, which is named BinWaves, consists on disagregating the full wave spectra into its different energy “bins” which are characterized by monochromatic wave trains represented by a frequency and a direction.
Based on the linearity of the waves in deep waters, every directional spectra can be later on aggregated by supperposing all its different constituents at each location and time.
Methodology
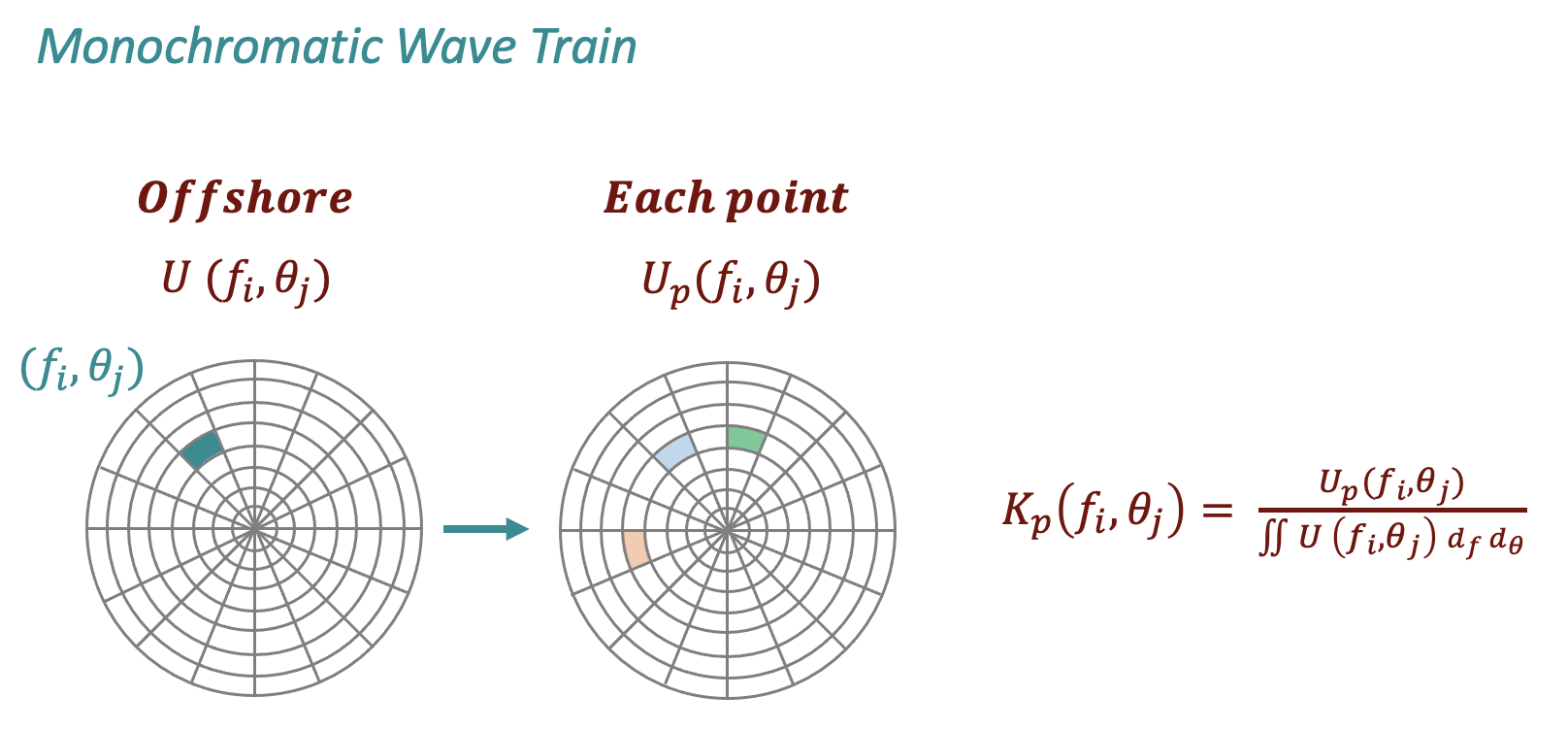
These are the steps followed in the BinWaves methodology
Divide the full energy spectra into its individual energy “bins”. The spectra, characterized by 29 frequencies and 24 directions, results in 696 monochromatic wave trains.
Numerically run the 696 monochromatic cases with unitary significant wave height.
Post process the 696 simulations to obtain the propagation coefficients (Kp) of each energy bin at every location.
The propagation coefficients nearshore, as exemplified in the sketch below at each point of the grid, can be characterized by more than 1 different directional component due to the interaction with the bathymetry along the propagation.
SWAN
Individual “bins” have been numerically simulated using SWAN (Simulating WAves Nearshore), which is a third-generation open source wave model developed at Delft University of Technology.
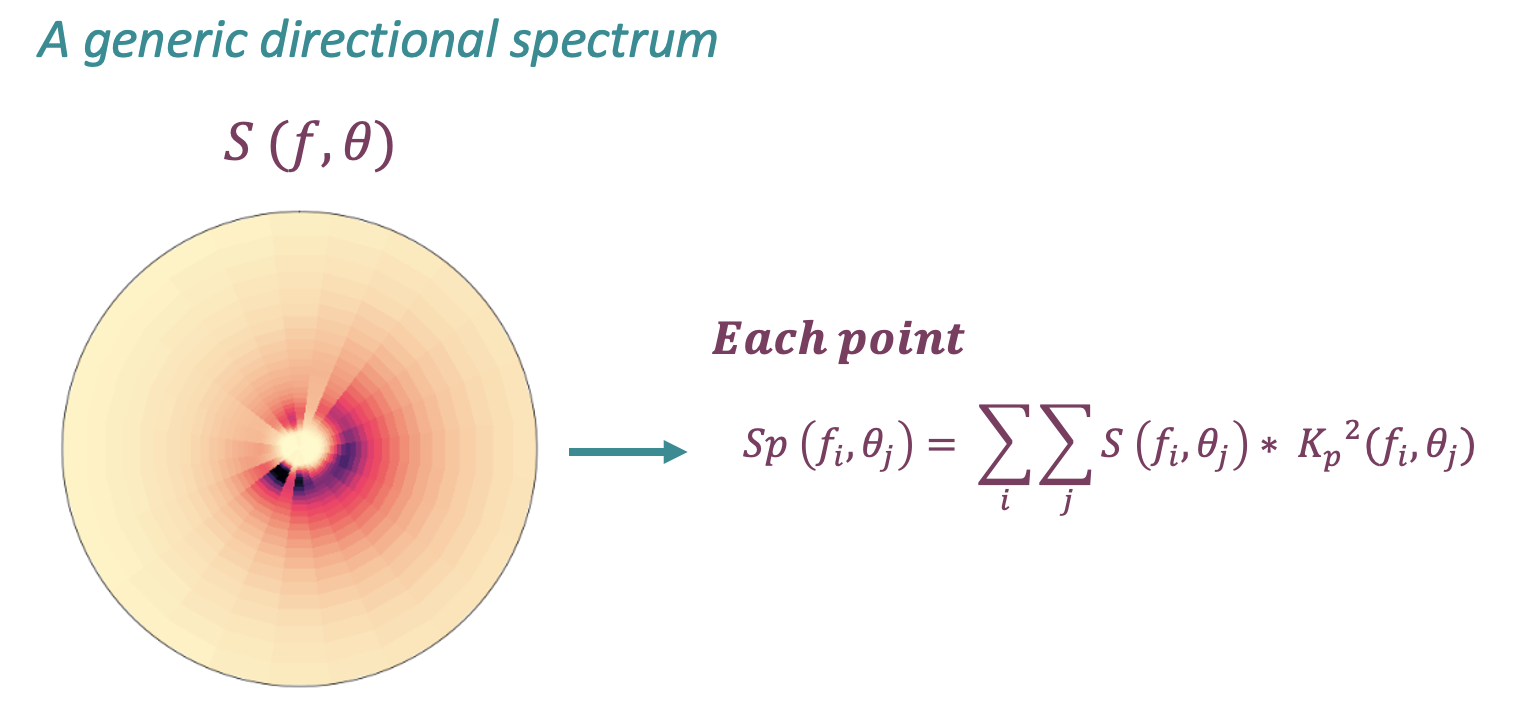
Reconstruction methodology
Once the outputs of the BinWaves methodology have been obtained at each point of the grid, for every generic wave spectra, the propagated spectra can be reconstructed by supperposing all of its components following the equation below.
Database and site parameters#
# database
p_data = '/media/administrador/HD2/SamoaTonga/data'
site = 'Samoa'
p_site = op.join(p_data, site)
# site bathymetry
p_bathy = op.join(p_site, 'bathymetry')
# deliverable folder
p_deliv = op.join(p_site, 'd05_swell_inundation_forecast')
p_spec = op.join(p_deliv, 'spec')
p_swan = op.join(p_deliv, 'swan')
# SWAN grid depth and parameters
p_grid_depth = op.join(p_bathy, 'grid300_depth.pkl')
p_grid_param = op.join(p_bathy, 'grid300_params.pkl')
# launch SWAN
launch_swan = False
# superpoint
deg_sup = 15
p_superpoint = op.join(p_spec, 'super_point_superposition_{0}_.nc'.format(deg_sup))
# output files
name = 'binwaves'
p_swan_subset = op.join(p_swan, name, 'subset.nc')
p_swan_output = op.join(p_swan, name, 'out_main_binwaves.nc')
p_swan_input_spec = op.join(p_swan, name, 'input_spec.nc')
# Load superpoint
sp = xr.open_dataset(p_superpoint)
dirs = sp.dir.values
freqs = sp.freq.values
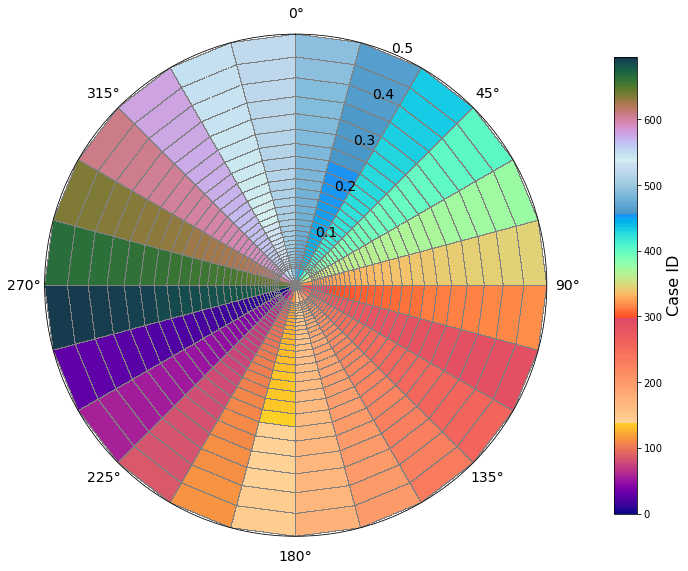
A number of 696 cases need to be run, corresponding to all the combinations of the spectra disctretized in 24 directions and 29 frequencies.
Below is an example of one of the monochromatic cases, with a freq ~0.3 (T = 3.3s) and Dir = 75
Plot_grid_cells(sp, pos=[25, 12]);
Obtain the parameters of the monochromatic cases
# each SWAN case is defined by 5 wave parameters: hs, tp, dir, spr, gamma
vns = ['hs','per','dir','spr','gamma']
gamma = 50 # waves gamma
spr = 3 # waves directional spread
# initialize case variables
cases = np.full([len(sp.freq) * len(sp.dir), len(vns)], np.nan)
cases_id = np.full([len(sp.freq), len(sp.dir)], np.nan)
c = 0 # case counter
# generate one case for each superpoint direction and frequency values
for ix_d, d in enumerate(sp.dir.values):
for ix_f, f in enumerate(sp.freq.values):
# hs switch
if 1 / f > 5:
hs = 1
else:
hs = 0.1
# set case parameters
cases[c, :] = [hs, np.round(1/f,4), d, spr, gamma]
# set case ID
cases_id[ix_f, ix_d] = c
c+=1
# cases DataFrame
df = pd.DataFrame(cases, columns = vns)
# plot cases ID
Plot_grid_cases(sp, cases_id.astype('int'));
SWAN Project#
These are the different steps that need to be followed to run SWAN:
Create project
Define bathymetry and parameters
Define simulation parameters
Build cases
Run cases
Create project
name = 'binwaves'
sp = SwanProject(p_swan, name)
if not op.isdir(op.join(p_swan, name)):
os.makedirs(op.join(p_swan, name))
Load bathymetry and its parameters
swan_grid = pk.load(open(p_grid_param, "rb"))
swan_depth = pk.load(open(p_grid_depth, "rb"))
Define bathymetry
# generate project main mesh
main_mesh = SwanMesh()
# bathymetry grid description
main_mesh.dg = swan_grid
# bathymetry depth
main_mesh.depth = swan_depth
# computational grid description
main_mesh.cg = main_mesh.dg.copy()
# set project main mesh
sp.set_main_mesh(main_mesh)
Define SWAN cases
input_params = {
'set_level': 0,
'set_convention': 'NAUTICAL',
'coords_mode': 'SPHERICAL',
'coords_projection': 'CCM',
'boundw_jonswap': gamma,
'boundw_period': 'PEAK',
'boundn_mode': 'CLOSED',
'wind': False,
'only_wind': False,
'physics': [
'FRICTION JONSWAP',
'BREAKING',
],
'numerics':[
'OFF QUAD',
],
'output_variables': [
'HSIGN', 'TM02', 'DIR', 'TPS', 'DSPR',
'PTHSIGN', 'PTRTP', 'PTDIR', 'PTDSPR',
],
'output_variables_points': [
'HSIGN', 'TM02', 'DIR', 'TPS', 'DSPR',
'PTHSIGN', 'PTRTP', 'PTDIR', 'PTDSPR',
],
}
sp.set_params(input_params)
main_mesh.dg
{'xpc': 187.08083499999807,
'ypc': -14.149377999999974,
'alpc': 0,
'xlenc': 1.5989999999924862,
'ylenc': 0.8370000000000317,
'mxc': 533,
'myc': 279,
'dxinp': 0.002999999999985903,
'dyinp': 0.0030000000000001137}
sp.params
{'set_level': 0,
'set_maxerr': None,
'set_cdcap': None,
'set_convention': 'NAUTICAL',
'coords_mode': 'SPHERICAL',
'coords_projection': 'CCM',
'cgrid_mdc': 72,
'cgrid_flow': 0.03,
'cgrid_fhigh': 1.0,
'boundw_jonswap': 50,
'boundw_period': 'PEAK',
'boundn_mode': 'CLOSED',
'physics': ['FRICTION JONSWAP', 'BREAKING'],
'numerics': ['OFF QUAD'],
'wind_deltinp': None,
'level_deltinp': None,
'compute_deltc': None,
'output_deltt': None,
'output_variables': ['HSIGN',
'TM02',
'DIR',
'TPS',
'DSPR',
'PTHSIGN',
'PTRTP',
'PTDIR',
'PTDSPR'],
'output_time_ini_specout': None,
'output_time_ini_block': None,
'output_time_ini_table': None,
'output_points_x': [],
'output_points_y': [],
'output_variables_points': ['HSIGN',
'TM02',
'DIR',
'TPS',
'DSPR',
'PTHSIGN',
'PTRTP',
'PTDIR',
'PTDSPR'],
'output_spec_deltt': None,
'output_spec': False,
'output_points_spec': False,
'wind': False,
'only_wind': False}
Swan wrap
sw = SwanWrap_STAT(sp)
Build and launch cases
subset = df.to_xarray().rename({'index':'case'})
# store a copy of the subset
subset.to_netcdf(p_swan_subset)
subset
<xarray.Dataset>
Dimensions: (case: 696)
Coordinates:
* case (case) int64 0 1 2 3 4 5 6 7 8 ... 688 689 690 691 692 693 694 695
Data variables:
hs (case) float64 1.0 1.0 1.0 1.0 1.0 1.0 ... 0.1 0.1 0.1 0.1 0.1 0.1
per (case) float64 28.57 25.97 23.61 21.47 ... 2.637 2.397 2.179 1.981
dir (case) float64 262.5 262.5 262.5 262.5 ... 277.5 277.5 277.5 277.5
spr (case) float64 3.0 3.0 3.0 3.0 3.0 3.0 ... 3.0 3.0 3.0 3.0 3.0 3.0
gamma (case) float64 50.0 50.0 50.0 50.0 50.0 ... 50.0 50.0 50.0 50.0- case: 696
- case(case)int640 1 2 3 4 5 ... 691 692 693 694 695
array([ 0, 1, 2, ..., 693, 694, 695])
- hs(case)float641.0 1.0 1.0 1.0 ... 0.1 0.1 0.1 0.1
array([1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, ... 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1]) - per(case)float6428.57 25.97 23.61 ... 2.179 1.981
array([28.5714, 25.974 , 23.6128, 21.4661, 19.5147, 17.7406, 16.1278, 14.6617, 13.3288, 12.1171, 11.0155, 10.0141, 9.1037, 8.2761, 7.5237, 6.8398, 6.218 , 5.6527, 5.1388, 4.6717, 4.247 , 3.8609, 3.5099, 3.1908, 2.9007, 2.637 , 2.3973, 2.1794, 1.9812, 28.5714, 25.974 , 23.6128, 21.4661, 19.5147, 17.7406, 16.1278, 14.6617, 13.3288, 12.1171, 11.0155, 10.0141, 9.1037, 8.2761, 7.5237, 6.8398, 6.218 , 5.6527, 5.1388, 4.6717, 4.247 , 3.8609, 3.5099, 3.1908, 2.9007, 2.637 , 2.3973, 2.1794, 1.9812, 28.5714, 25.974 , 23.6128, 21.4661, 19.5147, 17.7406, 16.1278, 14.6617, 13.3288, 12.1171, 11.0155, 10.0141, 9.1037, 8.2761, 7.5237, 6.8398, 6.218 , 5.6527, 5.1388, 4.6717, 4.247 , 3.8609, 3.5099, 3.1908, 2.9007, 2.637 , 2.3973, 2.1794, 1.9812, 28.5714, 25.974 , 23.6128, 21.4661, 19.5147, 17.7406, 16.1278, 14.6617, 13.3288, 12.1171, 11.0155, 10.0141, 9.1037, 8.2761, 7.5237, 6.8398, 6.218 , 5.6527, 5.1388, 4.6717, 4.247 , 3.8609, 3.5099, 3.1908, 2.9007, 2.637 , 2.3973, 2.1794, 1.9812, 28.5714, 25.974 , 23.6128, 21.4661, 19.5147, 17.7406, 16.1278, 14.6617, 13.3288, 12.1171, 11.0155, 10.0141, 9.1037, 8.2761, 7.5237, 6.8398, 6.218 , 5.6527, 5.1388, 4.6717, 4.247 , 3.8609, 3.5099, 3.1908, ... 12.1171, 11.0155, 10.0141, 9.1037, 8.2761, 7.5237, 6.8398, 6.218 , 5.6527, 5.1388, 4.6717, 4.247 , 3.8609, 3.5099, 3.1908, 2.9007, 2.637 , 2.3973, 2.1794, 1.9812, 28.5714, 25.974 , 23.6128, 21.4661, 19.5147, 17.7406, 16.1278, 14.6617, 13.3288, 12.1171, 11.0155, 10.0141, 9.1037, 8.2761, 7.5237, 6.8398, 6.218 , 5.6527, 5.1388, 4.6717, 4.247 , 3.8609, 3.5099, 3.1908, 2.9007, 2.637 , 2.3973, 2.1794, 1.9812, 28.5714, 25.974 , 23.6128, 21.4661, 19.5147, 17.7406, 16.1278, 14.6617, 13.3288, 12.1171, 11.0155, 10.0141, 9.1037, 8.2761, 7.5237, 6.8398, 6.218 , 5.6527, 5.1388, 4.6717, 4.247 , 3.8609, 3.5099, 3.1908, 2.9007, 2.637 , 2.3973, 2.1794, 1.9812, 28.5714, 25.974 , 23.6128, 21.4661, 19.5147, 17.7406, 16.1278, 14.6617, 13.3288, 12.1171, 11.0155, 10.0141, 9.1037, 8.2761, 7.5237, 6.8398, 6.218 , 5.6527, 5.1388, 4.6717, 4.247 , 3.8609, 3.5099, 3.1908, 2.9007, 2.637 , 2.3973, 2.1794, 1.9812, 28.5714, 25.974 , 23.6128, 21.4661, 19.5147, 17.7406, 16.1278, 14.6617, 13.3288, 12.1171, 11.0155, 10.0141, 9.1037, 8.2761, 7.5237, 6.8398, 6.218 , 5.6527, 5.1388, 4.6717, 4.247 , 3.8609, 3.5099, 3.1908, 2.9007, 2.637 , 2.3973, 2.1794, 1.9812]) - dir(case)float64262.5 262.5 262.5 ... 277.5 277.5
array([262.5, 262.5, 262.5, 262.5, 262.5, 262.5, 262.5, 262.5, 262.5, 262.5, 262.5, 262.5, 262.5, 262.5, 262.5, 262.5, 262.5, 262.5, 262.5, 262.5, 262.5, 262.5, 262.5, 262.5, 262.5, 262.5, 262.5, 262.5, 262.5, 247.5, 247.5, 247.5, 247.5, 247.5, 247.5, 247.5, 247.5, 247.5, 247.5, 247.5, 247.5, 247.5, 247.5, 247.5, 247.5, 247.5, 247.5, 247.5, 247.5, 247.5, 247.5, 247.5, 247.5, 247.5, 247.5, 247.5, 247.5, 247.5, 232.5, 232.5, 232.5, 232.5, 232.5, 232.5, 232.5, 232.5, 232.5, 232.5, 232.5, 232.5, 232.5, 232.5, 232.5, 232.5, 232.5, 232.5, 232.5, 232.5, 232.5, 232.5, 232.5, 232.5, 232.5, 232.5, 232.5, 232.5, 232.5, 217.5, 217.5, 217.5, 217.5, 217.5, 217.5, 217.5, 217.5, 217.5, 217.5, 217.5, 217.5, 217.5, 217.5, 217.5, 217.5, 217.5, 217.5, 217.5, 217.5, 217.5, 217.5, 217.5, 217.5, 217.5, 217.5, 217.5, 217.5, 217.5, 202.5, 202.5, 202.5, 202.5, 202.5, 202.5, 202.5, 202.5, 202.5, 202.5, 202.5, 202.5, 202.5, 202.5, 202.5, 202.5, 202.5, 202.5, 202.5, 202.5, 202.5, 202.5, 202.5, 202.5, 202.5, 202.5, 202.5, 202.5, 202.5, 187.5, 187.5, 187.5, 187.5, 187.5, 187.5, 187.5, 187.5, 187.5, 187.5, 187.5, 187.5, 187.5, 187.5, 187.5, 187.5, 187.5, 187.5, 187.5, 187.5, 187.5, 187.5, 187.5, 187.5, 187.5, 187.5, 187.5, 187.5, 187.5, 172.5, 172.5, 172.5, 172.5, 172.5, 172.5, ... 352.5, 352.5, 352.5, 352.5, 352.5, 352.5, 352.5, 352.5, 352.5, 352.5, 352.5, 352.5, 352.5, 352.5, 352.5, 352.5, 352.5, 352.5, 352.5, 352.5, 352.5, 352.5, 352.5, 352.5, 352.5, 352.5, 352.5, 352.5, 352.5, 337.5, 337.5, 337.5, 337.5, 337.5, 337.5, 337.5, 337.5, 337.5, 337.5, 337.5, 337.5, 337.5, 337.5, 337.5, 337.5, 337.5, 337.5, 337.5, 337.5, 337.5, 337.5, 337.5, 337.5, 337.5, 337.5, 337.5, 337.5, 337.5, 322.5, 322.5, 322.5, 322.5, 322.5, 322.5, 322.5, 322.5, 322.5, 322.5, 322.5, 322.5, 322.5, 322.5, 322.5, 322.5, 322.5, 322.5, 322.5, 322.5, 322.5, 322.5, 322.5, 322.5, 322.5, 322.5, 322.5, 322.5, 322.5, 307.5, 307.5, 307.5, 307.5, 307.5, 307.5, 307.5, 307.5, 307.5, 307.5, 307.5, 307.5, 307.5, 307.5, 307.5, 307.5, 307.5, 307.5, 307.5, 307.5, 307.5, 307.5, 307.5, 307.5, 307.5, 307.5, 307.5, 307.5, 307.5, 292.5, 292.5, 292.5, 292.5, 292.5, 292.5, 292.5, 292.5, 292.5, 292.5, 292.5, 292.5, 292.5, 292.5, 292.5, 292.5, 292.5, 292.5, 292.5, 292.5, 292.5, 292.5, 292.5, 292.5, 292.5, 292.5, 292.5, 292.5, 292.5, 277.5, 277.5, 277.5, 277.5, 277.5, 277.5, 277.5, 277.5, 277.5, 277.5, 277.5, 277.5, 277.5, 277.5, 277.5, 277.5, 277.5, 277.5, 277.5, 277.5, 277.5, 277.5, 277.5, 277.5, 277.5, 277.5, 277.5, 277.5, 277.5]) - spr(case)float643.0 3.0 3.0 3.0 ... 3.0 3.0 3.0 3.0
array([3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., ... 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3., 3.]) - gamma(case)float6450.0 50.0 50.0 ... 50.0 50.0 50.0
array([50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., ... 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50., 50.])
# store SWAN input spectra for later use
sp_input = np.full([len(subset.case), len(freqs), len(dirs)], 0.0)
for case in subset.case:
inp = subset.sel(case = case)
k = 1.1
f = 1/inp.per.values
d = inp.dir.values
i = np.argmin(np.abs(freqs-f))
j = np.argmin(np.abs(dirs-d))
sp_input[case, i, j] = k
input_spec = xr.Dataset(
{
'hs': (['case','freq','dir'], sp_input),
},
coords = {
'case': subset.case,
'dir': dirs,
'freq': freqs
}
)
input_spec.to_netcdf(p_swan_input_spec)
input_spec
<xarray.Dataset>
Dimensions: (case: 696, dir: 24, freq: 29)
Coordinates:
* case (case) int64 0 1 2 3 4 5 6 7 8 ... 688 689 690 691 692 693 694 695
* dir (dir) float32 262.5 247.5 232.5 217.5 ... 322.5 307.5 292.5 277.5
* freq (freq) float32 0.035 0.0385 0.04235 ... 0.4171 0.4589 0.5047
Data variables:
hs (case, freq, dir) float64 1.1 0.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 1.1- case: 696
- dir: 24
- freq: 29
- case(case)int640 1 2 3 4 5 ... 691 692 693 694 695
array([ 0, 1, 2, ..., 693, 694, 695])
- dir(dir)float32262.5 247.5 232.5 ... 292.5 277.5
array([262.5, 247.5, 232.5, 217.5, 202.5, 187.5, 172.5, 157.5, 142.5, 127.5, 112.5, 97.5, 82.5, 67.5, 52.5, 37.5, 22.5, 7.5, 352.5, 337.5, 322.5, 307.5, 292.5, 277.5], dtype=float32) - freq(freq)float320.035 0.0385 ... 0.4589 0.5047
array([0.035 , 0.0385 , 0.04235 , 0.046585, 0.051244, 0.056368, 0.062005, 0.068205, 0.075026, 0.082528, 0.090781, 0.099859, 0.109845, 0.12083 , 0.132912, 0.146204, 0.160824, 0.176907, 0.194597, 0.214057, 0.235463, 0.259009, 0.28491 , 0.313401, 0.344741, 0.379215, 0.417136, 0.45885 , 0.504735], dtype=float32)
- hs(case, freq, dir)float641.1 0.0 0.0 0.0 ... 0.0 0.0 0.0 1.1
array([[[1.1, 0. , 0. , ..., 0. , 0. , 0. ], [0. , 0. , 0. , ..., 0. , 0. , 0. ], [0. , 0. , 0. , ..., 0. , 0. , 0. ], ..., [0. , 0. , 0. , ..., 0. , 0. , 0. ], [0. , 0. , 0. , ..., 0. , 0. , 0. ], [0. , 0. , 0. , ..., 0. , 0. , 0. ]], [[0. , 0. , 0. , ..., 0. , 0. , 0. ], [1.1, 0. , 0. , ..., 0. , 0. , 0. ], [0. , 0. , 0. , ..., 0. , 0. , 0. ], ..., [0. , 0. , 0. , ..., 0. , 0. , 0. ], [0. , 0. , 0. , ..., 0. , 0. , 0. ], [0. , 0. , 0. , ..., 0. , 0. , 0. ]], [[0. , 0. , 0. , ..., 0. , 0. , 0. ], [0. , 0. , 0. , ..., 0. , 0. , 0. ], [1.1, 0. , 0. , ..., 0. , 0. , 0. ], ..., ... ..., [0. , 0. , 0. , ..., 0. , 0. , 1.1], [0. , 0. , 0. , ..., 0. , 0. , 0. ], [0. , 0. , 0. , ..., 0. , 0. , 0. ]], [[0. , 0. , 0. , ..., 0. , 0. , 0. ], [0. , 0. , 0. , ..., 0. , 0. , 0. ], [0. , 0. , 0. , ..., 0. , 0. , 0. ], ..., [0. , 0. , 0. , ..., 0. , 0. , 0. ], [0. , 0. , 0. , ..., 0. , 0. , 1.1], [0. , 0. , 0. , ..., 0. , 0. , 0. ]], [[0. , 0. , 0. , ..., 0. , 0. , 0. ], [0. , 0. , 0. , ..., 0. , 0. , 0. ], [0. , 0. , 0. , ..., 0. , 0. , 0. ], ..., [0. , 0. , 0. , ..., 0. , 0. , 0. ], [0. , 0. , 0. , ..., 0. , 0. , 0. ], [0. , 0. , 0. , ..., 0. , 0. , 1.1]]])
if launch_swan:
# build and launch swan cases
sw.build_cases(df)
sw.run_cases()
Data extraction#
Once the SWAN propagations have been run, we have to extract the results in the different points of the grid
# extract swan cases output
memory_safe = False
if memory_safe:
# This output extraction uses cases chunks to avoid memory problems
chunk_size = 100
# prepare a folder to store the chunks
p_chunks = op.join(p_swan, name, 'extract_output')
if not op.isdir(p_chunks): os.makedirs(p_chunks)
# generate chunks of cases to extract
slices = [[chunk_size*x, chunk_size*(x+1)] for x in range(int(np.ceil(len(df)/chunk_size)))]
slices[-1][1] = len(df)
# extract and store each chunk
for c_ini, c_end in slices:
out_chk = sw.extract_output(case_ini=c_ini, case_end=c_end)
out_chk.to_netcdf(op.join(p_chunks, 'output_chunk_{0}_{1}.nc'.format(c_ini, c_end)))
# combine all chunks into one output file
out_sea_mm_sim = xr.open_mfdataset(op.join(p_chunks, '*.nc'))
# clean temporal folder
shutil.rmtree(p_chunks)
else:
# load entire cases output at once
out_sea_mm_sim = sw.extract_output()
SWAN output postprocess: fix 0.1 Hs cases and add a partition number counter
# Rescale simulations of hs=0.01 multiplying by 10
pos_01 = np.where(subset['hs'] < 1)[0]
# get base Hs values
Hsig = out_sea_mm_sim.Hsig.values
Hs_part = out_sea_mm_sim.Hs_part.values
# fix Hs values
for c in range(len(out_sea_mm_sim.case)):
if np.isin(c, pos_01):
Hsig[c,:,:] = Hsig[c,:,:] * 10
Hs_part[c,:,:,:] = Hs_part[c,:,:,:] * 10
# override Hsig and Hs_part variables
out_sea_mm_sim['Hsig'] = (('case','lat','lon'), Hsig)
out_sea_mm_sim['Hs_part'] = (('case','partition','lat','lon'), Hs_part)
# Calculate number of partitions at each location
matrix_count = np.full(np.shape(out_sea_mm_sim.Hs_part), 0.0)
matrix_count[np.where(out_sea_mm_sim.Hs_part > 0)] = 1
cont = np.sum(matrix_count, axis=1)
cont[np.where(cont == 0)] = np.nan
# add counter to SWAN output
out_sea_mm_sim['count_parts'] = (('case','lat','lon'), cont)
Store SWAN output
out_sea_mm_sim
<xarray.Dataset>
Dimensions: (case: 696, lat: 279, lon: 533, partition: 10)
Coordinates:
* lon (lon) float64 187.1 187.1 187.1 187.1 ... 188.7 188.7 188.7
* lat (lat) float64 -14.15 -14.15 -14.14 ... -13.32 -13.32 -13.32
* case (case) int64 0 1 2 3 4 5 6 7 ... 689 690 691 692 693 694 695
Dimensions without coordinates: partition
Data variables:
Tp (case, lat, lon) float32 27.76 27.76 27.76 ... 1.939 1.939
Dir (case, lat, lon) float32 262.5 262.5 262.5 ... 277.4 277.4
Tm02 (case, lat, lon) float32 26.62 26.62 26.62 ... 1.81 1.81 1.81
Hsig (case, lat, lon) float32 0.9978 0.9978 0.9978 ... 1.039 1.039
Dspr (case, lat, lon) float32 3.837 3.849 3.861 ... 3.057 3.058
Hs_part (case, partition, lat, lon) float32 0.9932 0.9932 ... 0.0 0.0
Tp_part (case, partition, lat, lon) float32 27.81 27.81 ... 0.0 0.0
Dir_part (case, partition, lat, lon) float32 262.5 262.5 ... 0.0 0.0
Dspr_part (case, partition, lat, lon) float32 3.831 3.842 ... 0.0 0.0
count_parts (case, lat, lon) float64 1.0 1.0 1.0 1.0 ... 1.0 1.0 1.0 1.0- case: 696
- lat: 279
- lon: 533
- partition: 10
- lon(lon)float64187.1 187.1 187.1 ... 188.7 188.7
array([187.080835, 187.083835, 187.086835, ..., 188.670835, 188.673835, 188.676835]) - lat(lat)float64-14.15 -14.15 ... -13.32 -13.32
array([-14.149378, -14.146378, -14.143378, ..., -13.321378, -13.318378, -13.315378]) - case(case)int640 1 2 3 4 5 ... 691 692 693 694 695
array([ 0, 1, 2, ..., 693, 694, 695])
- Tp(case, lat, lon)float3227.76 27.76 27.76 ... 1.939 1.939
- units :
- s
- description :
- Waves Peak Period
array([[[27.763784 , 27.763788 , 27.76379 , ..., 27.774172 , 27.773745 , 27.773355 ], [27.763771 , 27.763773 , 27.763775 , ..., 27.759596 , 27.759758 , 27.760036 ], [27.763767 , 27.763773 , 27.763773 , ..., 27.746183 , 27.746996 , 27.747774 ], ..., [27.763676 , 27.763676 , 27.763676 , ..., 27.610516 , 27.60782 , 27.606045 ], [27.763676 , 27.763678 , 27.763676 , ..., 27.677097 , 27.673895 , 27.671291 ], [27.763676 , 27.763676 , 27.763676 , ..., 27.74151 , 27.739017 , 27.736517 ]], [[25.242155 , 25.242159 , 25.24216 , ..., 25.245134 , 25.244875 , 25.244616 ], [25.242151 , 25.24215 , 25.242151 , ..., 25.237385 , 25.237389 , 25.237438 ], [25.242157 , 25.242155 , 25.242159 , ..., 25.228308 , 25.228552 , 25.228844 ], ... [ 2.132025 , 2.1320286, 2.1320322, ..., 2.132108 , 2.1321082, 2.1321082], [ 2.132025 , 2.1320283, 2.132031 , ..., 2.1320734, 2.1320736, 2.1320739], [ 2.132025 , 2.1320252, 2.1320252, ..., 2.132039 , 2.132039 , 2.132039 ]], [[ 1.938941 , 1.9389491, 1.9389575, ..., 1.9415294, 1.9415315, 1.9415339], [ 1.938941 , 1.9389493, 1.9389575, ..., 1.9415228, 1.9415245, 1.9415263], [ 1.938941 , 1.9389493, 1.9389576, ..., 1.9415084, 1.9415092, 1.9415113], ..., [ 1.938941 , 1.9389493, 1.9389576, ..., 1.939119 , 1.9391191, 1.939119 ], [ 1.938941 , 1.9389483, 1.9389548, ..., 1.9390424, 1.9390423, 1.9390424], [ 1.938941 , 1.938941 , 1.938941 , ..., 1.9389641, 1.9389642, 1.9389642]]], dtype=float32) - Dir(case, lat, lon)float32262.5 262.5 262.5 ... 277.4 277.4
- units :
- º
- description :
- Waves Direction
array([[[262.5022 , 262.5022 , 262.5022 , ..., 263.4983 , 263.50113, 263.50128], [262.5019 , 262.50134, 262.50085, ..., 263.39133, 263.3837 , 263.37018], [262.50153, 262.5009 , 262.5002 , ..., 263.13666, 263.12363, 263.10776], ..., [262.5 , 262.4993 , 262.4986 , ..., 266.49588, 266.4562 , 266.4134 ], [262.5 , 262.4993 , 262.4986 , ..., 266.71387, 266.68414, 266.64822], [262.5 , 262.49936, 262.49872, ..., 266.89032, 266.875 , 266.85233]], [[262.50137, 262.50134, 262.50134, ..., 263.46033, 263.46988, 263.47656], [262.5012 , 262.5006 , 262.5001 , ..., 263.47498, 263.4718 , 263.46143], [262.50098, 262.50027, 262.4996 , ..., 263.33005, 263.31598, 263.2985 ], ... [277.49997, 277.4993 , 277.4986 , ..., 277.42764, 277.4274 , 277.42648], [277.49997, 277.49942, 277.49896, ..., 277.42645, 277.42633, 277.42587], [277.49997, 277.49982, 277.49966, ..., 277.4244 , 277.42432, 277.4241 ]], [[277.49997, 277.49936, 277.49872, ..., 276.88943, 276.85904, 276.82938], [277.49997, 277.4992 , 277.49847, ..., 276.741 , 276.7106 , 276.68402], [277.49997, 277.4992 , 277.49844, ..., 276.60046, 276.5731 , 276.55045], ..., [277.49997, 277.4993 , 277.4986 , ..., 277.46967, 277.46954, 277.46893], [277.49997, 277.4995 , 277.49927, ..., 277.45798, 277.45792, 277.45764], [277.49997, 277.49985, 277.49973, ..., 277.4442 , 277.44415, 277.44403]]], dtype=float32) - Tm02(case, lat, lon)float3226.62 26.62 26.62 ... 1.81 1.81
- units :
- s
- description :
- Waves Mean Period
array([[[26.615091 , 26.615103 , 26.615122 , ..., 26.673521 , 26.671772 , 26.669985 ], [26.615044 , 26.615055 , 26.615082 , ..., 26.607832 , 26.608025 , 26.608389 ], [26.615086 , 26.61509 , 26.6151 , ..., 26.52907 , 26.53145 , 26.533827 ], ..., [26.614388 , 26.614397 , 26.614405 , ..., 25.71602 , 25.70033 , 25.689886 ], [26.614388 , 26.614399 , 26.614403 , ..., 26.138002 , 26.120188 , 26.105978 ], [26.614388 , 26.614399 , 26.614399 , ..., 26.555832 , 26.542461 , 26.529102 ]], [[24.227144 , 24.227156 , 24.227165 , ..., 24.268713 , 24.267994 , 24.267197 ], [24.22713 , 24.227154 , 24.22716 , ..., 24.237188 , 24.237072 , 24.236847 ], [24.227165 , 24.227182 , 24.227194 , ..., 24.193926 , 24.194452 , 24.194954 ], ... [ 1.995612 , 1.9974326, 1.999296 , ..., 2.0256755, 2.0256777, 2.025603 ], [ 1.995612 , 1.9972543, 1.9986941, ..., 2.012039 , 2.0120401, 2.0120432], [ 1.995612 , 1.9956168, 1.9956214, ..., 1.9968123, 1.9968139, 1.9968162]], [[ 1.8087884, 1.8117365, 1.814613 , ..., 1.9546384, 1.9546504, 1.9546627], [ 1.8087884, 1.8117439, 1.814657 , ..., 1.9547102, 1.954719 , 1.9547273], [ 1.8087884, 1.8117443, 1.8146579, ..., 1.9547249, 1.9547288, 1.954738 ], ..., [ 1.8087884, 1.8117145, 1.8146931, ..., 1.8507851, 1.8507862, 1.8506061], [ 1.8087884, 1.8114305, 1.813726 , ..., 1.8319062, 1.8319074, 1.8319088], [ 1.8087884, 1.8087962, 1.8088034, ..., 1.8097959, 1.8097967, 1.809798 ]]], dtype=float32) - Hsig(case, lat, lon)float320.9978 0.9978 ... 1.039 1.039
array([[[0.99775475, 0.99775505, 0.9977554 , ..., 1.0371958 , 1.0371983 , 1.037116 ], [0.9977489 , 0.99774504, 0.9977396 , ..., 1.0227605 , 1.0229003 , 1.0228506 ], [0.99776834, 0.99776566, 0.997764 , ..., 1.0002964 , 1.0005742 , 1.0007328 ], ..., [0.99752986, 0.99753004, 0.99753 , ..., 0.32569492, 0.32479507, 0.32424843], [0.9975298 , 0.99753004, 0.99753046, ..., 0.35803965, 0.35660192, 0.35552418], [0.9975298 , 0.9975329 , 0.9975361 , ..., 0.40329906, 0.40157238, 0.39990714]], [[1.0040985 , 1.0040969 , 1.0040951 , ..., 1.0388212 , 1.0391244 , 1.0393332 ], [1.0040998 , 1.0040948 , 1.0040872 , ..., 1.0338826 , 1.0339731 , 1.03381 ], [1.0041155 , 1.0041134 , 1.004111 , ..., 1.0214972 , 1.021374 , 1.0210984 ], ... [1.035128 , 1.0347626 , 1.0343869 , ..., 1.0331864 , 1.0331942 , 1.033218 ], [1.035128 , 1.0348024 , 1.0345196 , ..., 1.0360132 , 1.0360185 , 1.0360296 ], [1.035128 , 1.0351361 , 1.0351441 , ..., 1.038998 , 1.0390036 , 1.0390112 ]], [[1.0364414 , 1.0355508 , 1.0346711 , ..., 0.82857704, 0.82618356, 0.82382095], [1.0364414 , 1.0355577 , 1.0346811 , ..., 0.81842595, 0.8158816 , 0.8135074 ], [1.0364414 , 1.035558 , 1.0346822 , ..., 0.80660105, 0.8040404 , 0.8018934 ], ..., [1.0364414 , 1.0355681 , 1.0346731 , ..., 1.0250657 , 1.0250698 , 1.0251039 ], [1.0364414 , 1.0356579 , 1.0349731 , ..., 1.0319153 , 1.0319185 , 1.0319252 ], [1.0364414 , 1.0364482 , 1.0364549 , ..., 1.0392941 , 1.0392975 , 1.0393021 ]]], dtype=float32) - Dspr(case, lat, lon)float323.837 3.849 3.861 ... 3.057 3.058
- units :
- º
- description :
- Waves Directional Spread
array([[[3.8370092, 3.8486695, 3.8607 , ..., 4.7143025, 4.719612 , 4.719985 ], [3.817428 , 3.8202457, 3.828739 , ..., 4.7595754, 4.742153 , 4.7150908], [3.8841958, 3.8790543, 3.877389 , ..., 4.6418157, 4.60083 , 4.561449 ], ..., [2.9696581, 2.9698558, 2.9702513, ..., 6.665827 , 6.7594547, 6.8845105], [2.9695263, 2.9698558, 2.9699216, ..., 5.9970517, 6.084895 , 6.201661 ], [2.9693286, 2.9696581, 2.970185 , ..., 5.3249416, 5.376358 , 5.470881 ]], [[3.5363941, 3.5417018, 3.5479395, ..., 4.7757454, 4.788675 , 4.796391 ], [3.541039 , 3.540818 , 3.54358 , ..., 4.8967986, 4.8877196, 4.867702 ], [3.5990272, 3.595056 , 3.5923336, ..., 4.829404 , 4.7982674, 4.7646294], ... [2.9692626, 2.9695263, 2.969724 , ..., 3.0882635, 3.0893402, 3.0949724], [2.9692626, 2.9696581, 2.9699216, ..., 3.0873127, 3.0876932, 3.0903537], [2.9692626, 2.9693286, 2.9696581, ..., 3.0873127, 3.0873764, 3.088327 ]], [[2.969197 , 2.9693947, 2.9693947, ..., 2.772211 , 2.7494607, 2.726664 ], [2.969197 , 2.9696581, 2.969724 , ..., 2.6632793, 2.6389227, 2.618378 ], [2.969197 , 2.9694605, 2.969724 , ..., 2.5643134, 2.5423963, 2.5241678], ..., [2.969197 , 2.9693947, 2.9699874, ..., 3.05264 , 3.053153 , 3.0566115], [2.969197 , 2.9694605, 2.9697897, ..., 3.0544984, 3.0546265, 3.0559714], [2.969197 , 2.9693947, 2.9695263, ..., 3.0571237, 3.0571878, 3.0578277]]], dtype=float32) - Hs_part(case, partition, lat, lon)float320.9932 0.9932 0.9932 ... 0.0 0.0
array([[[[0.9932059 , 0.99320614, 0.9932064 , ..., 1.0167929 , 1.0166545 , 1.0164739 ], [0.99320036, 0.9931965 , 0.99319065, ..., 1.0008736 , 1.0011889 , 1.0014409 ], [0.99321985, 0.99321723, 0.99321544, ..., 0.9746652 , 0.975603 , 0.97645223], ..., [0.99298376, 0.99298394, 0.992984 , ..., 0.31948817, 0.31836942, 0.31759274], [0.99298376, 0.99298406, 0.99298435, ..., 0.35221756, 0.35061944, 0.34934184], [0.99298376, 0.99298686, 0.99299 , ..., 0.39766094, 0.3958523 , 0.39406374]], [[0. , 0. , 0. , ..., 0.17842141, 0.17925148, 0.17982922], [0. , 0. , 0. , ..., 0.18707481, 0.18608303, 0.18438713], [0. , 0. , 0. , ..., 0.20516098, 0.20190978, 0.19845845], ... [0. , 0. , 0. , ..., 0. , 0. , 0. ], [0. , 0. , 0. , ..., 0. , 0. , 0. ], [0. , 0. , 0. , ..., 0. , 0. , 0. ]], [[0. , 0. , 0. , ..., 0. , 0. , 0. ], [0. , 0. , 0. , ..., 0. , 0. , 0. ], [0. , 0. , 0. , ..., 0. , 0. , 0. ], ..., [0. , 0. , 0. , ..., 0. , 0. , 0. ], [0. , 0. , 0. , ..., 0. , 0. , 0. ], [0. , 0. , 0. , ..., 0. , 0. , 0. ]]]], dtype=float32) - Tp_part(case, partition, lat, lon)float3227.81 27.81 27.81 ... 0.0 0.0 0.0
- units :
- s
- description :
- Partition of Waves Peak Period
array([[[[27.81195 , 27.811954 , 27.811958 , ..., 27.848438 , 27.848307 , 27.848017 ], [27.81194 , 27.811941 , 27.811943 , ..., 27.834389 , 27.83426 , 27.83394 ], [27.811937 , 27.811937 , 27.81194 , ..., 27.801767 , 27.802578 , 27.803293 ], ..., [27.811832 , 27.811832 , 27.811832 , ..., 27.643194 , 27.640322 , 27.638462 ], [27.811832 , 27.811832 , 27.811832 , ..., 27.716621 , 27.71329 , 27.71062 ], [27.811832 , 27.811832 , 27.811832 , ..., 27.787521 , 27.785023 , 27.78251 ]], [[ 0. , 0. , 0. , ..., 27.466322 , 27.465303 , 27.465258 ], [ 0. , 0. , 0. , ..., 27.46511 , 27.465212 , 27.466293 ], [ 0. , 0. , 0. , ..., 27.632309 , 27.629503 , 27.627216 ], ... [ 0. , 0. , 0. , ..., 0. , 0. , 0. ], [ 0. , 0. , 0. , ..., 0. , 0. , 0. ], [ 0. , 0. , 0. , ..., 0. , 0. , 0. ]], [[ 0. , 0. , 0. , ..., 0. , 0. , 0. ], [ 0. , 0. , 0. , ..., 0. , 0. , 0. ], [ 0. , 0. , 0. , ..., 0. , 0. , 0. ], ..., [ 0. , 0. , 0. , ..., 0. , 0. , 0. ], [ 0. , 0. , 0. , ..., 0. , 0. , 0. ], [ 0. , 0. , 0. , ..., 0. , 0. , 0. ]]]], dtype=float32) - Dir_part(case, partition, lat, lon)float32262.5 262.5 262.5 ... 0.0 0.0 0.0
- units :
- º
- description :
- Partition of Waves Direction
array([[[[262.5022 , 262.5022 , 262.5022 , ..., 263.03802, 263.03784, 263.03516], [262.5019 , 262.50134, 262.50085, ..., 262.8711 , 262.86896, 262.86353], [262.50153, 262.50085, 262.50018, ..., 262.47064, 262.47906, 262.48523], ..., [262.5 , 262.49927, 262.4986 , ..., 267.45844, 267.4572 , 267.45612], [262.5 , 262.49933, 262.4986 , ..., 267.46765, 267.46655, 267.4655 ], [262.5 , 262.49933, 262.49872, ..., 267.4732 , 267.47247, 267.47174]], [[ 0. , 0. , 0. , ..., 278.623 , 278.5752 , 278.5664 ], [ 0. , 0. , 0. , ..., 278.54645, 278.5453 , 278.5758 ], [ 0. , 0. , 0. , ..., 278.44263, 278.4406 , 278.44373], ... [ 0. , 0. , 0. , ..., 0. , 0. , 0. ], [ 0. , 0. , 0. , ..., 0. , 0. , 0. ], [ 0. , 0. , 0. , ..., 0. , 0. , 0. ]], [[ 0. , 0. , 0. , ..., 0. , 0. , 0. ], [ 0. , 0. , 0. , ..., 0. , 0. , 0. ], [ 0. , 0. , 0. , ..., 0. , 0. , 0. ], ..., [ 0. , 0. , 0. , ..., 0. , 0. , 0. ], [ 0. , 0. , 0. , ..., 0. , 0. , 0. ], [ 0. , 0. , 0. , ..., 0. , 0. , 0. ]]]], dtype=float32) - Dspr_part(case, partition, lat, lon)float323.831 3.842 3.854 ... 0.0 0.0 0.0
- units :
- º
- description :
- Partition of directional spread
array([[[[ 3.830885 , 3.8421564, 3.8541567, ..., 3.8396091, 3.8528364, 3.8510075], [ 3.812145 , 3.8145566, 3.8226016, ..., 3.7766144, 3.7664976, 3.7432559], [ 3.8790545, 3.8737543, 3.8723395, ..., 3.2636068, 3.259167 , 3.2573054], ..., [ 2.9693286, 2.9700537, 2.9702513, ..., 1.4219912, 1.4390888, 1.4514092], [ 2.9693286, 2.9695263, 2.96979 , ..., 1.290864 , 1.3065335, 1.3202398], [ 2.9693286, 2.9699216, 2.9700537, ..., 1.1497608, 1.1617811, 1.1756771]], [[ 0. , 0. , 0. , ..., 5.7038016, 5.478887 , 5.4034047], [ 0. , 0. , 0. , ..., 5.346029 , 5.331222 , 5.3949604], [ 0. , 0. , 0. , ..., 5.086101 , 5.0800576, 5.075896 ], ... [ 0. , 0. , 0. , ..., 0. , 0. , 0. ], [ 0. , 0. , 0. , ..., 0. , 0. , 0. ], [ 0. , 0. , 0. , ..., 0. , 0. , 0. ]], [[ 0. , 0. , 0. , ..., 0. , 0. , 0. ], [ 0. , 0. , 0. , ..., 0. , 0. , 0. ], [ 0. , 0. , 0. , ..., 0. , 0. , 0. ], ..., [ 0. , 0. , 0. , ..., 0. , 0. , 0. ], [ 0. , 0. , 0. , ..., 0. , 0. , 0. ], [ 0. , 0. , 0. , ..., 0. , 0. , 0. ]]]], dtype=float32) - count_parts(case, lat, lon)float641.0 1.0 1.0 1.0 ... 1.0 1.0 1.0 1.0
array([[[1., 1., 1., ..., 2., 2., 2.], [1., 1., 1., ..., 2., 2., 2.], [1., 1., 1., ..., 2., 2., 2.], ..., [1., 1., 1., ..., 2., 2., 2.], [1., 1., 1., ..., 2., 2., 2.], [1., 1., 1., ..., 2., 2., 2.]], [[1., 1., 1., ..., 2., 2., 2.], [1., 1., 1., ..., 2., 2., 2.], [1., 1., 1., ..., 2., 2., 2.], ..., [1., 1., 1., ..., 2., 2., 2.], [1., 1., 1., ..., 2., 2., 2.], [1., 1., 1., ..., 2., 2., 2.]], [[1., 1., 1., ..., 2., 2., 2.], [1., 1., 1., ..., 2., 2., 2.], [1., 1., 1., ..., 2., 2., 2.], ..., ... ..., [1., 1., 1., ..., 1., 1., 1.], [1., 1., 1., ..., 1., 1., 1.], [1., 1., 1., ..., 1., 1., 1.]], [[1., 1., 1., ..., 1., 1., 1.], [1., 1., 1., ..., 1., 1., 1.], [1., 1., 1., ..., 1., 1., 1.], ..., [1., 1., 1., ..., 1., 1., 1.], [1., 1., 1., ..., 1., 1., 1.], [1., 1., 1., ..., 1., 1., 1.]], [[1., 1., 1., ..., 1., 1., 1.], [1., 1., 1., ..., 1., 1., 1.], [1., 1., 1., ..., 1., 1., 1.], ..., [1., 1., 1., ..., 1., 1., 1.], [1., 1., 1., ..., 1., 1., 1.], [1., 1., 1., ..., 1., 1., 1.]]])
# store output to netcdf
# store_xarray_max_compression(out_sea_mm_sim, p_swan_output)
out_sea_mm_sim.to_netcdf(p_swan_output)
out_sea_mm_sim = xr.open_dataset(p_swan_output)
print(out_sea_mm_sim)
<xarray.Dataset>
Dimensions: (case: 696, lat: 279, lon: 533, partition: 10)
Coordinates:
* lon (lon) float64 187.1 187.1 187.1 187.1 ... 188.7 188.7 188.7
* lat (lat) float64 -14.15 -14.15 -14.14 ... -13.32 -13.32 -13.32
* case (case) int64 0 1 2 3 4 5 6 7 ... 689 690 691 692 693 694 695
Dimensions without coordinates: partition
Data variables:
Tp (case, lat, lon) float32 ...
Dir (case, lat, lon) float32 ...
Tm02 (case, lat, lon) float32 ...
Hsig (case, lat, lon) float32 ...
Dspr (case, lat, lon) float32 ...
Hs_part (case, partition, lat, lon) float32 ...
Tp_part (case, partition, lat, lon) float32 ...
Dir_part (case, partition, lat, lon) float32 ...
Dspr_part (case, partition, lat, lon) float32 ...
count_parts (case, lat, lon) float64 ...
Plotting#
Individual cases#
Note
Once the cases have been run and outputs have been extracted, here we plot some results.
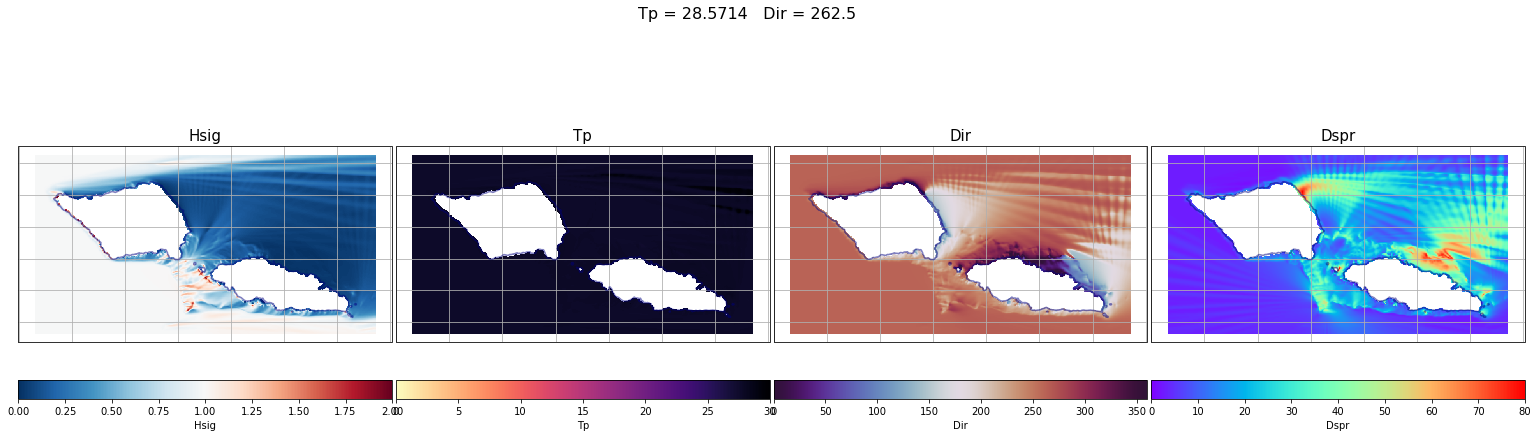
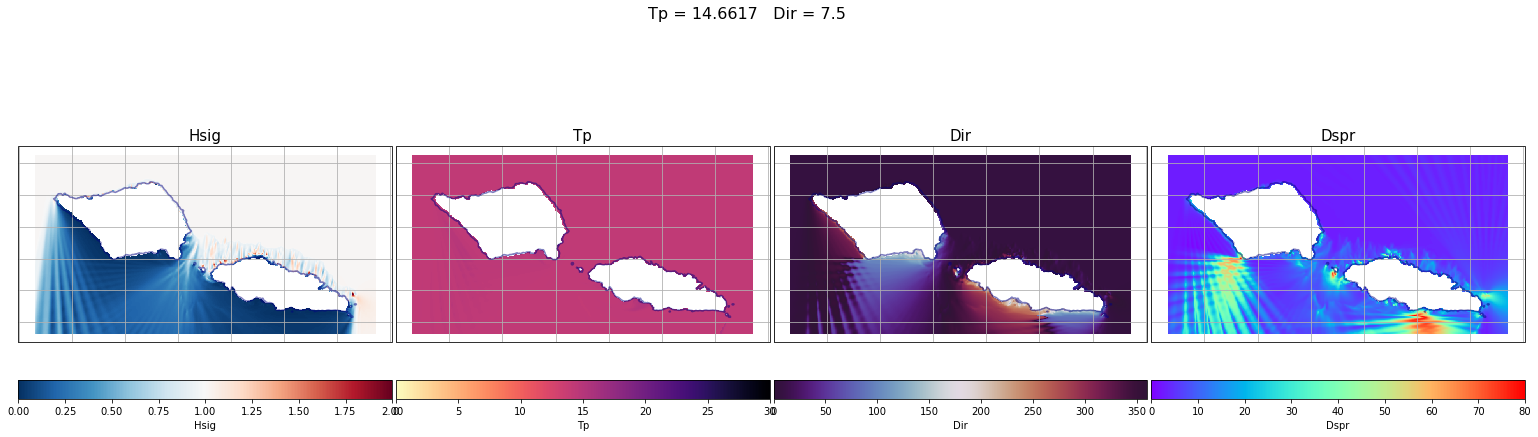
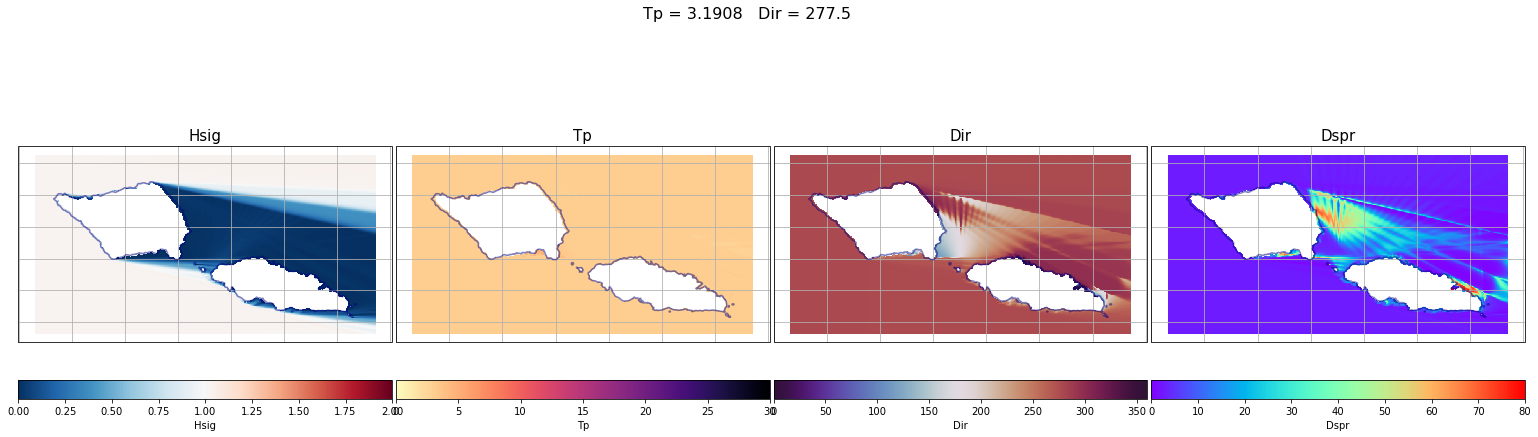
First, some individual cases have been plot. From left to right the different variables are: Hs (which corresponds to the propagation coefficient Kp, since we are running unitary wave heights), Tp, Dir and Dspr for the bulk parameters of the simulation, although partitions will be used to determine the propagation coefficients for each case.
case = 0
lon_s = 184
lat_s = -21
Plot_swan_case(out_sea_mm_sim, subset, case);
case = 500
lon_s = 184
lat_s = -21
Plot_swan_case(out_sea_mm_sim, subset, case);
case = 690
lon_s = 184
lat_s = -21
Plot_swan_case(out_sea_mm_sim, subset, case);
All cases#
Note
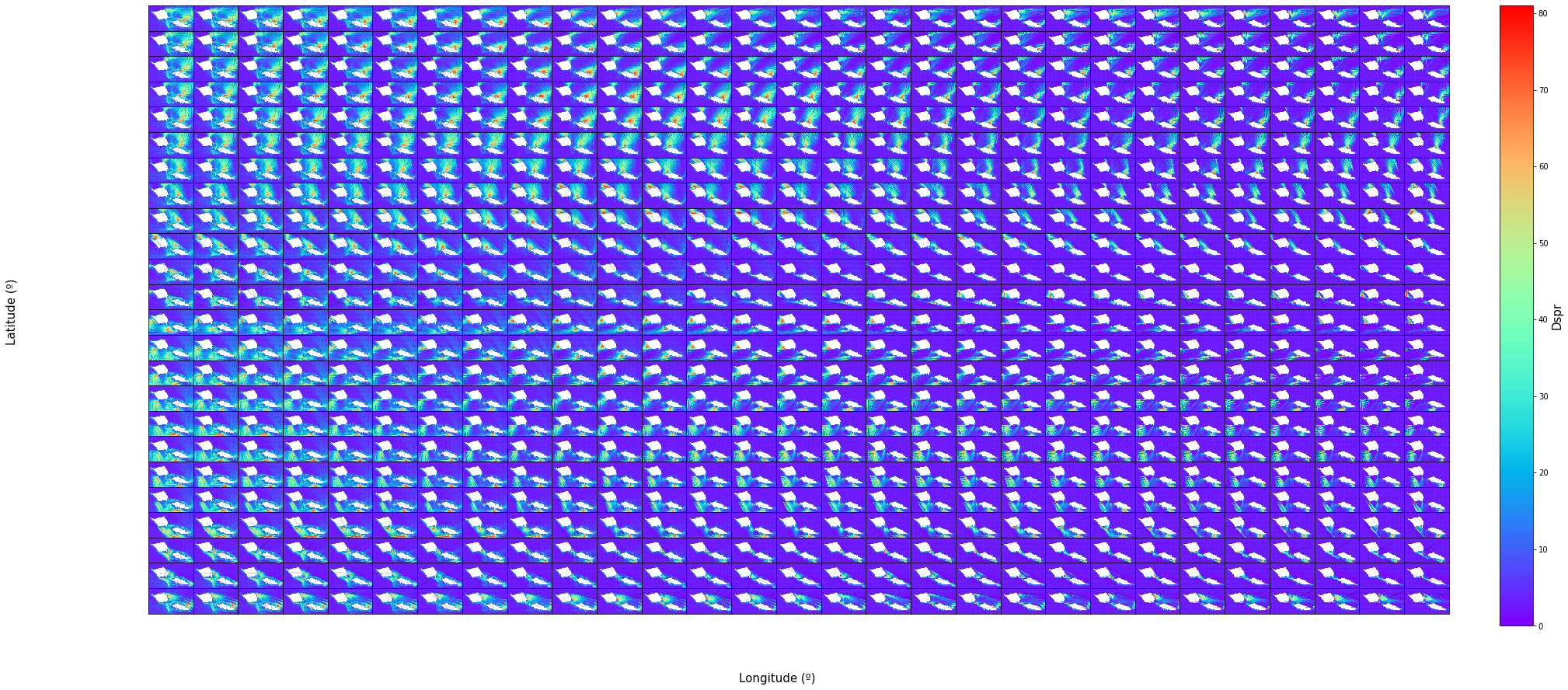
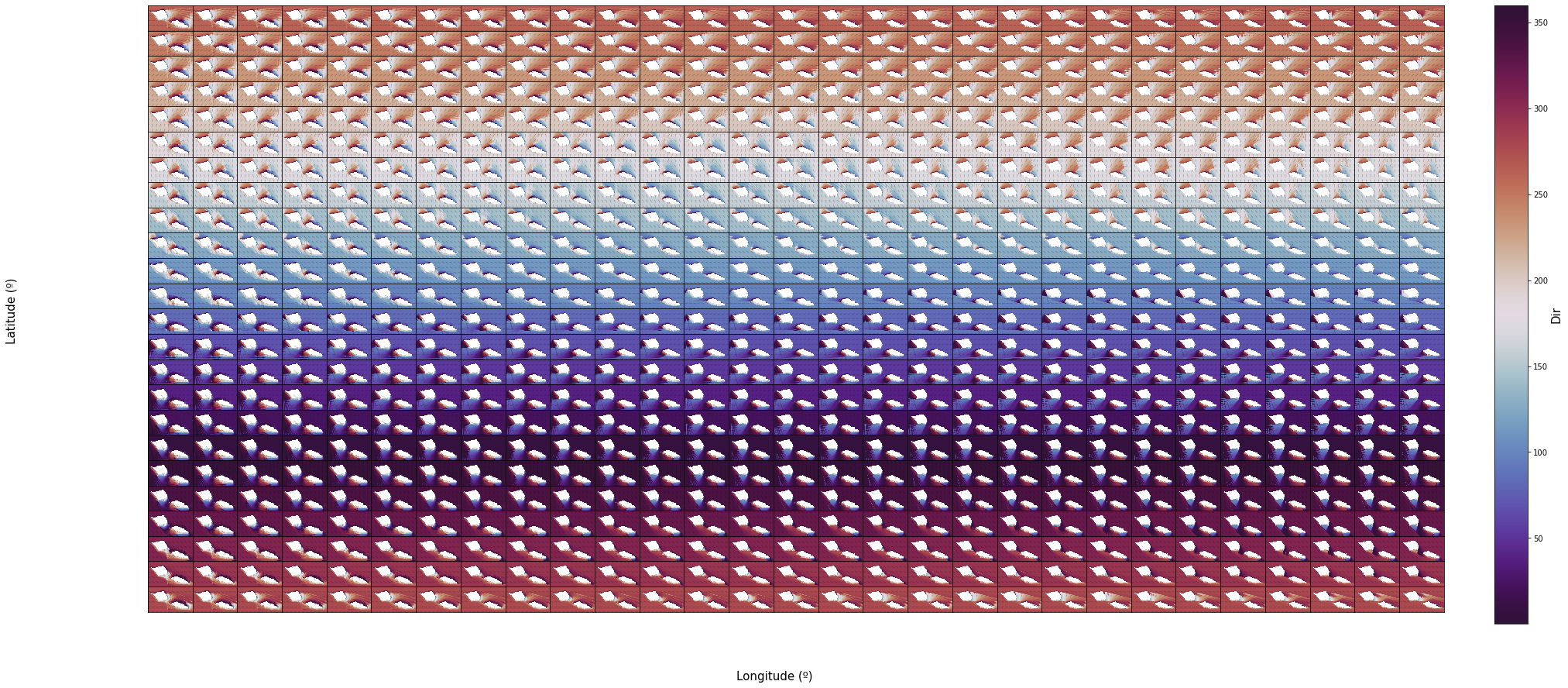
The grids bellow contain the same results but for all of the 696 cases ordered by Dir in rows and Tp in columns.
Hsig
nc = 29
nr = 24
n_cases = nc * nr
vars_ = ['Hsig']
scatter_maps(
out_sea_mm_sim,
n_cases = n_cases,
n_cols = nc, n_rows = nr,
var_list = vars_,
figsize = [30, 0.6 * nr],
var_limits = {'Hsig': (0, 2)},
);

TPsmoo
nc = 29
nr = 24
n_cases = nc * nr
vars_ = ['Tp']
scatter_maps(
out_sea_mm_sim,
n_cases = n_cases,
n_cols = nc, n_rows = nr,
var_list = vars_,
figsize = [30, 0.6 * nr],
);
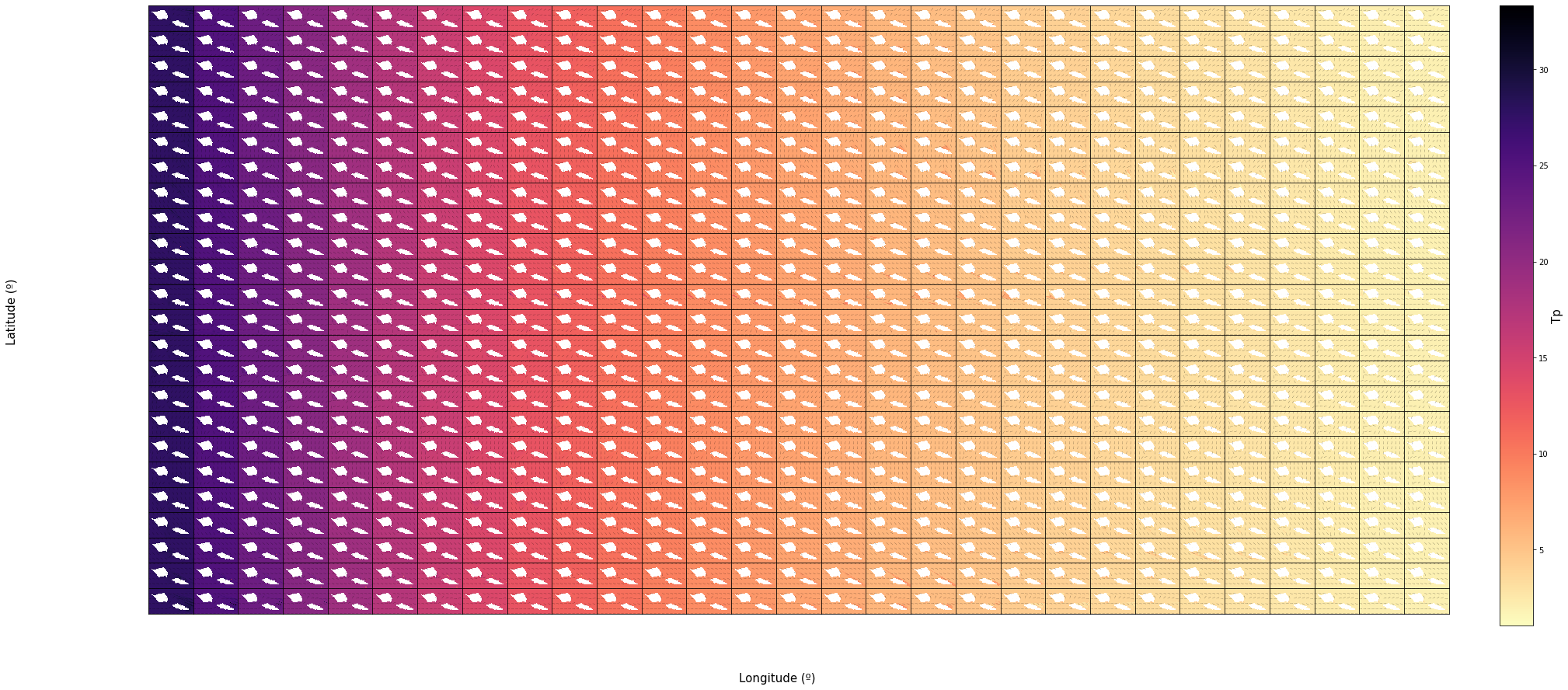
Dir
nc = 29
nr = 24
n_cases = nc * nr
vars_ = ['Dir']
scatter_maps(
out_sea_mm_sim,
n_cases = n_cases,
n_cols = nc, n_rows = nr,
var_list = vars_,
figsize = [30, 0.6 * nr],
);
Dspr
nc = 29
nr = 24
n_cases = nc * nr
vars_ = ['Dspr']
scatter_maps(
out_sea_mm_sim,
n_cases = n_cases,
n_cols = nc, n_rows = nr,
var_list = vars_,
figsize = [30, 0.6 * nr],
);