Tropical Cyclones
Contents
5. Tropical Cyclones#
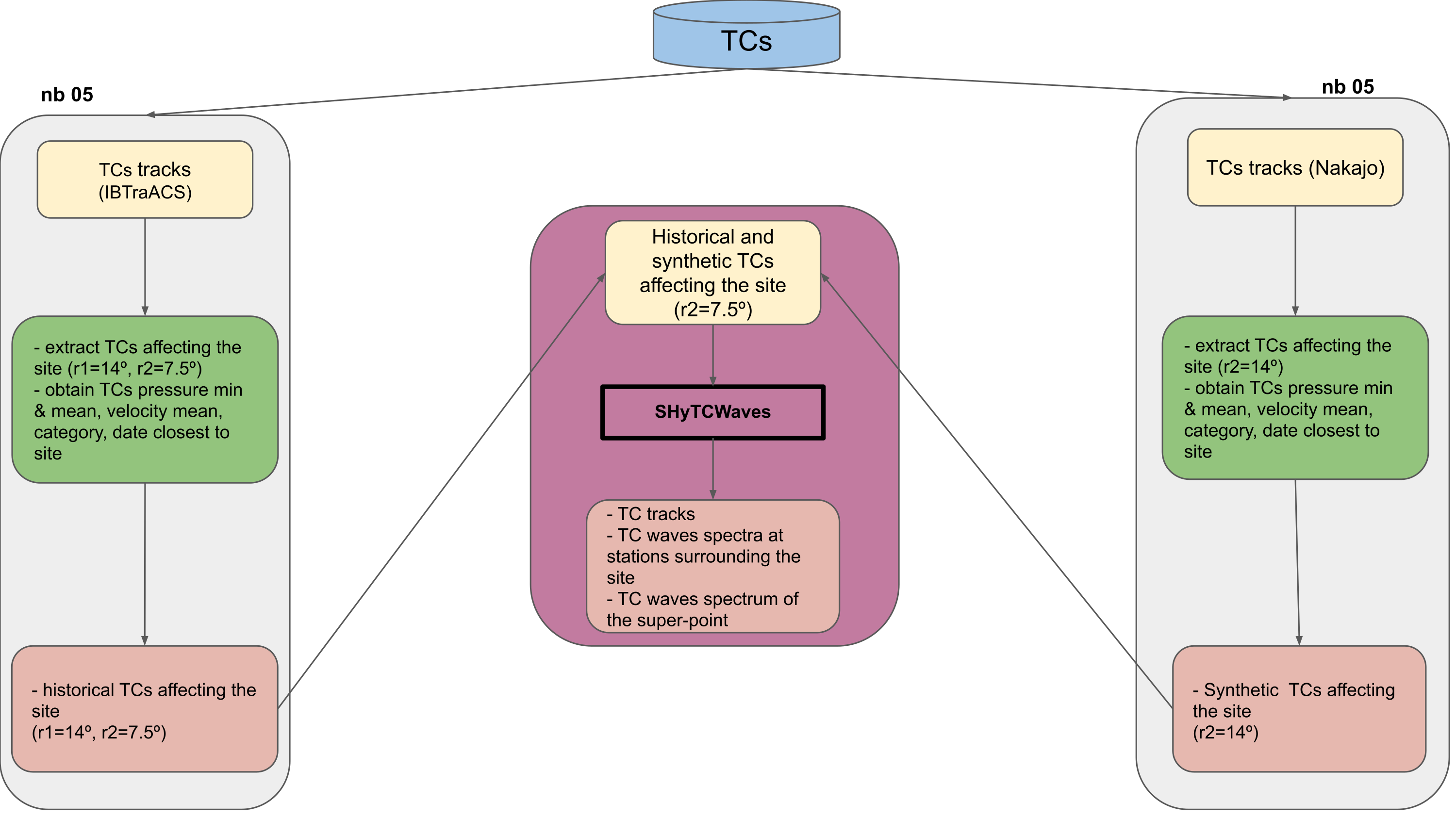
Selection of historical and synthetic Tropical Cyclones (TCs) affecting the site
inputs required:
Storm tracks records (ibtracks file)
Synthetic Storm tracks records from Nakajo database (Nakajo et al. 2014)
longitude and latitude of the site (location of the waves record)
radius for TCs selection (º)
in this notebook:
selection of TCs affecting the site for including in Daily Weather Type time series(14º around the site)
selection of TCs affecting the site for numerical simulation (7.5º around the site)
obtain TC category, minimum pressure, date when the TC track is closest to site
Workflow:
#!/usr/bin/env python
# -*- coding: utf-8 -*-
# common
import os
import os.path as op
import copy
# pip
import xarray as xr
import numpy as np
# DEV: override installed teslakit
import sys
sys.path.insert(0, op.join(os.path.abspath(''), '..', '..', '..'))
# teslakit
from bluemath.teslakit.tcs.tcs import Extract_Circle, filter_simultaneous_TCs
from bluemath.teslakit.io.aux import save_nc
from bluemath.teslakit.io.matlab import ReadNakajoMats
from bluemath.teslakit.plotting.tcs import Plot_TCs_HistoricalTracks, Plot_TCs_HistoricalTracks_Category
from bluemath.teslakit.plotting.tcs import Plot_TCs_Params_HISTvsSIM, Plot_TCs_Params_HISTvsSIM_histogram
5.1. Files and paths#
# project path
p_data = '/Users/albacid/Projects/TeslaKit.2.0_projects/SAMOA'
# output path
p_out = op.join(p_data,'TCs')
if not os.path.isdir(p_out): os.makedirs(p_out)
# SAMOA wave point longitude(0-360) and latitude
site = 'samoa'
pnt_lon = -172.07+360
pnt_lat = -13.76
# input data
tcs_file = op.join(p_data,'resources','IBTrACS.ALL.v04r00.nc')
tcs_path = op.join(p_data,'resources','Nakajo_tracks')
# output data
tcs_hist_r1_file = op.join(p_out,'TCs_hist_r1_' + site + '_raw.nc') # TCs crossing through r1
tcs_hist_r1_params_file = op.join(p_out,'TCs_hist_r1_params_' + site + '_raw.nc') # TCs parameters r1
tcs_hist_r1_params_file_sel = op.join(p_out,'TCs_hist_r1_params_' + site + '.nc') # TCs parameters r1
tcs_hist_r2_params_file = op.join(p_out,'TCs_hist_r2_params_' + site + '_raw.nc') # TCs parameters r2
tcs_hist_r2_params_file_sel = op.join(p_out,'TCs_hist_r2_params_' + site + '.nc') # TCs parameters r2
tcs_sim_r1_file = op.join(p_out,'TCs_r1_sim_' + site + '.nc') # TCs crossing through r1
tcs_sim_r1_params_file = op.join(p_out,'TCs_r1_sim_params_' + site + '.nc') # TCs parameters r1
5.2. Parameters#
# --------------------------------------
# load data and set parameters
TCs_wmo = xr.open_dataset(tcs_file) # noaa IBTrACS.ALL.v04r00.nc
# Load Nakajo simulations
#TCs_Nakajo = ReadNakajoMats(tcs_path)
# save netcdf with Nakajo sims
#TCs_Nakajo.to_netcdf(op.join(p_data,'resources','TCs_nakajo_all.nc'))
TCs_Nakajo = xr.open_dataset(op.join(p_data,'resources','TCs_nakajo_all.nc'))
# radius for TCs selection (º)
r1 = 14
r2 = 7.5
5.3. Historical TCs - extraction#
# --------------------------------------
# Select Historical TCs inside circle
# dictionary with needed variable names
d_vns = {
'longitude': 'lon',
'latitude': 'lat',
'time': 'time',
'pressure': 'wmo_pres',
}
# Select TCs that crosses a circular area R1
TCs_r1_tracks, TCs_r1_params = Extract_Circle(TCs_wmo, pnt_lon, pnt_lat, r1, d_vns)
TCs_r2_tracks, TCs_r2_params = Extract_Circle(TCs_wmo, pnt_lon, pnt_lat, r2, d_vns)
# store data
save_nc(TCs_r1_tracks, tcs_hist_r1_file)
save_nc(TCs_r1_params, tcs_hist_r1_params_file)
save_nc(TCs_r2_params, tcs_hist_r2_params_file)
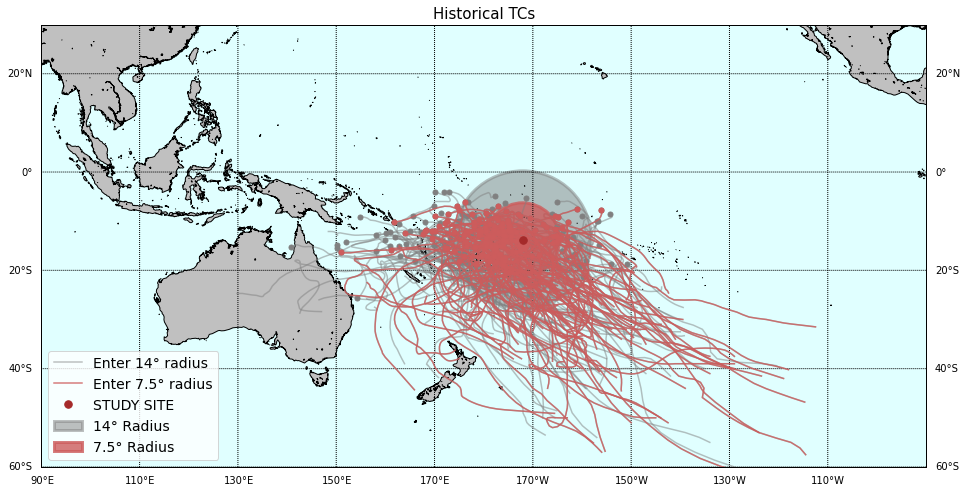
# Plot storm tracks world map (requires basemap)
lon1, lon2 = 90, 270
lat1, lat2 = -60, 30
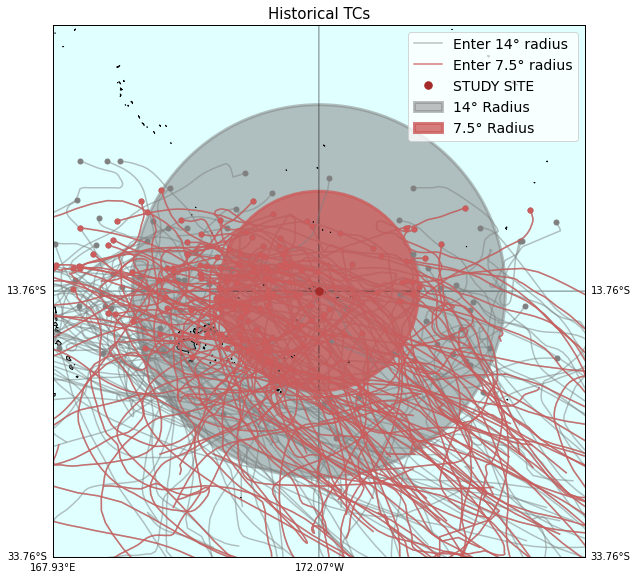
Plot_TCs_HistoricalTracks(
TCs_r1_tracks, TCs_r2_tracks,
lon1, lon2, lat1, lat2,
pnt_lon, pnt_lat, r1, r2,
);
# zoom map
r_zoom = 20
Plot_TCs_HistoricalTracks(
TCs_r1_tracks,TCs_r2_tracks,
pnt_lon-r_zoom, pnt_lon+r_zoom, pnt_lat-r_zoom, pnt_lat+r_zoom,
pnt_lon, pnt_lat, r1, r2
);
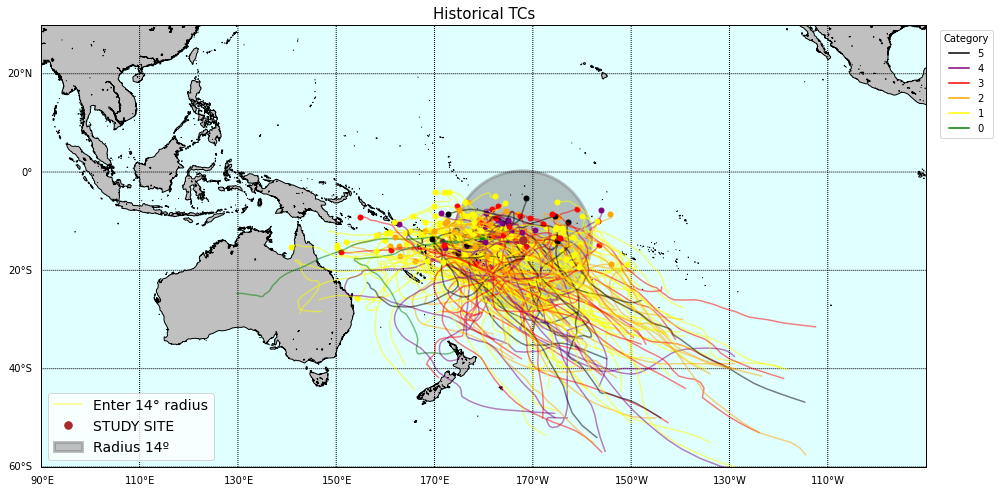
# Plot storm tracks category world map (requires basemap)
lon1, lon2 = 90, 270
lat1, lat2 = -60, 30
# r1
Plot_TCs_HistoricalTracks_Category(
TCs_r1_tracks, TCs_r1_params.category,
lon1, lon2, lat1, lat2,
pnt_lon, pnt_lat, r1,
);
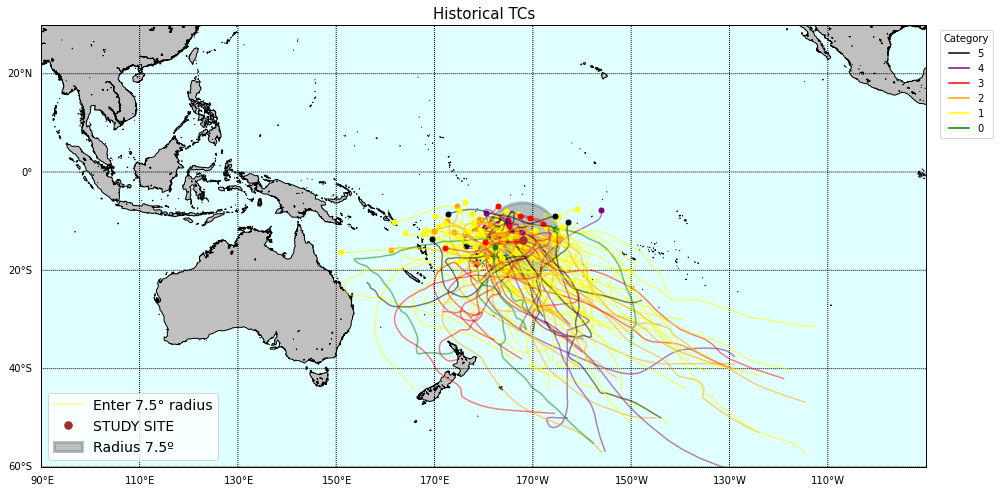
# r2
Plot_TCs_HistoricalTracks_Category(
TCs_r2_tracks, TCs_r2_params.category,
lon1, lon2, lat1, lat2,
pnt_lon, pnt_lat, r2,
);
5.3.1. Filter TCs that happen simultaneously#
# Keep TCs of greater category or longest track inside circle
# sort TCs by date
TCs_r1_params_sel = filter_simultaneous_TCs(TCs_r1_params)
TCs_r2_params_sel = filter_simultaneous_TCs(TCs_r2_params)
# save
save_nc(TCs_r1_params_sel, tcs_hist_r1_params_file_sel)
save_nc(TCs_r2_params_sel, tcs_hist_r2_params_file_sel)
5.4. Synthetic TCs - extraction#
# Get Nakajo TCs at the study area
# dictionary with needed variable names
d_vns = {
'longitude':'ylon_TC',
'latitude':'ylat_TC',
'time': 'yts',
'pressure':'yCPRES',
}
# Extract Nakajo TCs inside r1
TCs_r1_sim, TCs_r1_sim_params = Extract_Circle(TCs_Nakajo, pnt_lon, pnt_lat, r1, d_vns)
# Store Nakajo TCs
TCs_r1_sim.to_netcdf(tcs_sim_r1_file)
TCs_r1_sim_params.to_netcdf(tcs_sim_r1_params_file)
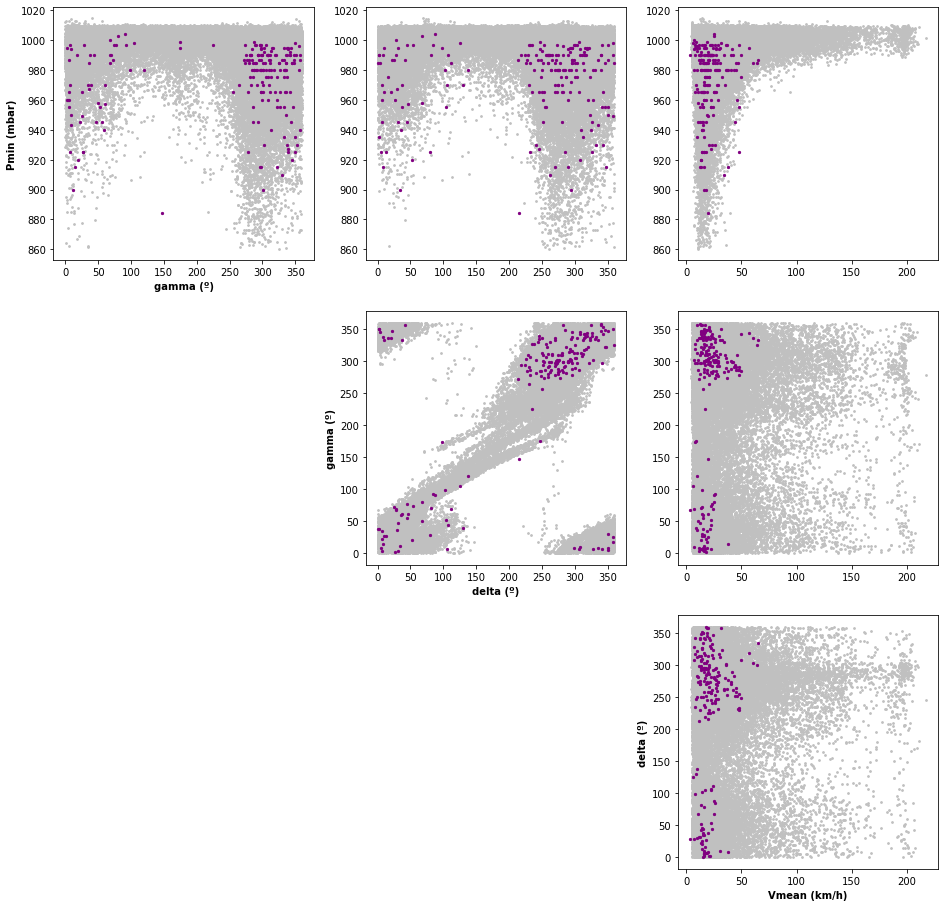
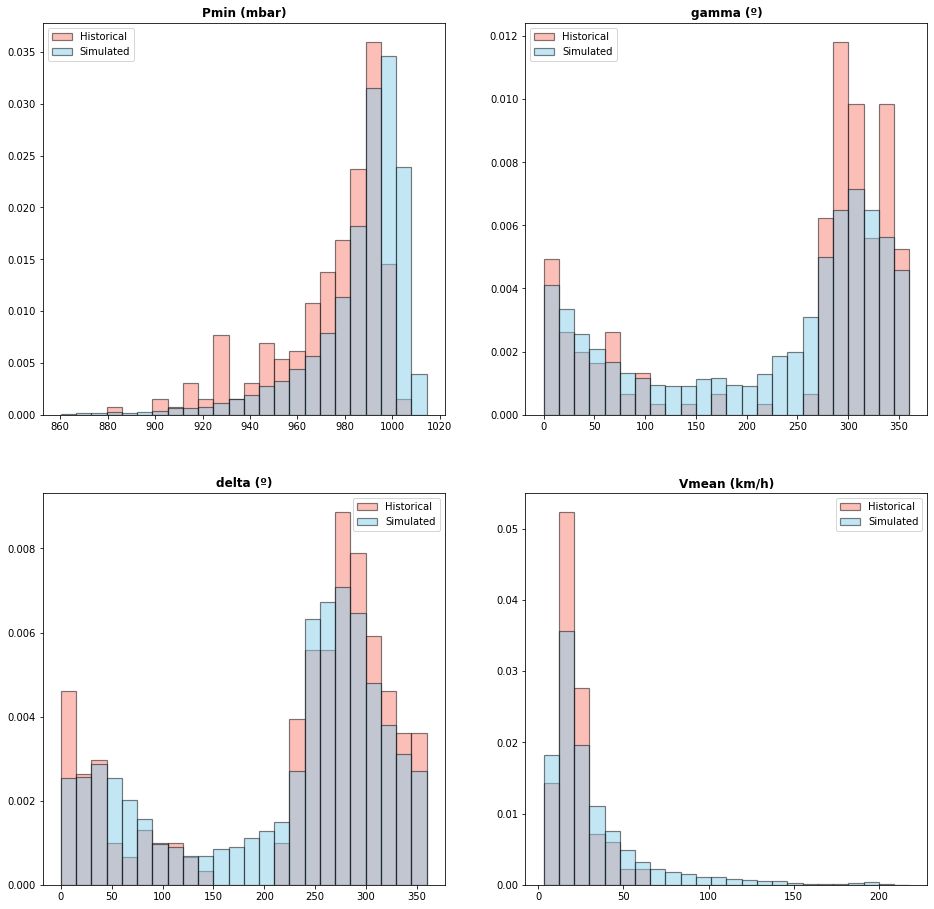
# Historical vs Nakajo Simulation parameters:
TCs_r1_hist_params = xr.open_dataset(tcs_hist_r1_params_file_sel) # historical TCs parameters inside radius 1
# scatter plot
Plot_TCs_Params_HISTvsSIM(TCs_r1_hist_params, TCs_r1_sim_params);
# histogram
Plot_TCs_Params_HISTvsSIM_histogram(TCs_r1_hist_params, TCs_r1_sim_params);