Flooding
Contents
Flooding#
Forecast
In this section we will produce the probabilistic rainfall assessment for a historical TC track with forecast information from JTWC issued some days prior to the TC arrival
from datetime import datetime
print("execution start: {0}".format(datetime.today().strftime('%Y-%m-%d %H:%M:%S')))
execution start: 2022-09-13 06:50:02
import os
import os.path as op
import sys
import glob
import time
from scipy.stats import qmc
import pandas as pd
import numpy as np
import xarray as xr
from sklearn.cluster import KMeans
import matplotlib.pyplot as plt
from matplotlib import gridspec
import cartopy.crs as ccrs
# raster tools
from rasterio.crs import CRS
import geopandas as gpd
import rioxarray
# bluemath
sys.path.insert(0, op.join(op.abspath(''), '..', '..'))
sys.path.insert(0, op.join(op.abspath(''), '..'))
from bluemath.teslakit.mda import MaxDiss_Simplified_NoThreshold
from bluemath.tc_warnings import download_warning
from bluemath.ddviewer.downloader.forecast import webscan_noaa_ncei, download_noaa
from bluemath.rainfall_forecast.rainfall_forecast import forecast_tracks, Reforecast_Track_Rain, \
tc_total_rain, tc_total_rain_site, Colors_basins, Plot_Rain_Hyetograph_Basins, \
Get_Analogues, Get_Flood_All_Basins, Plot_Rain_Flood_Points, \
Obtain_Basins
from bluemath.rainfall_forecast.tc_forecast import Load_TCs_Warning, Plot_tracks, Plot_Density_Forecast_Tracks
from bluemath.tc_forecast import get_info_TCs_warning, Load_TCs_Warning, Format_Tracks
from bluemath.tc_forecast import Plot_Tracks_Maxwinds, Plot_Tracks_Maxwinds_Density, Plot_Tracks_Maxwinds_zoom
# operational utils
from operational.util import clean_forecast_folder, read_config_args
Warning: ecCodes 2.21.0 or higher is recommended. You are running version 2.16.0
# aux functions
def Plot_Basins(GEO, ax=[], text=True, fill=True, figsize=[20,15]):
cols = Colors_basins(GEO)
if not ax:
fig = plt.figure(figsize=figsize)
gs = gridspec.GridSpec(1,1, wspace=0.001, hspace=0.001)
ax= plt.subplot(gs[0])
for b in np.arange(0,len(GEO)):
poly = GEO[b]
xs, ys = poly.exterior.xy
if text:
ax.text(np.double(poly.centroid.coords.xy[0][0]),np.double(poly.centroid.coords.xy[1][0]), str(b+1), fontsize=18, color='darkmagenta')#cols[b])
if fill:
ax.fill(xs, ys, alpha=0.5, fc=cols[b], linewidth=2, color=cols[b])
ax.plot(xs, ys, alpha=1, linewidth=3, color=cols[b])
ax.set_aspect('equal')
ax.set_xlabel('X UTM (m)', fontsize=20, fontweight='bold', color='navy')
ax.set_ylabel('Y_UTM (m)', fontsize=20, fontweight='bold', color='navy')
return fig
# hide warnings
import warnings
warnings.filterwarnings('ignore')
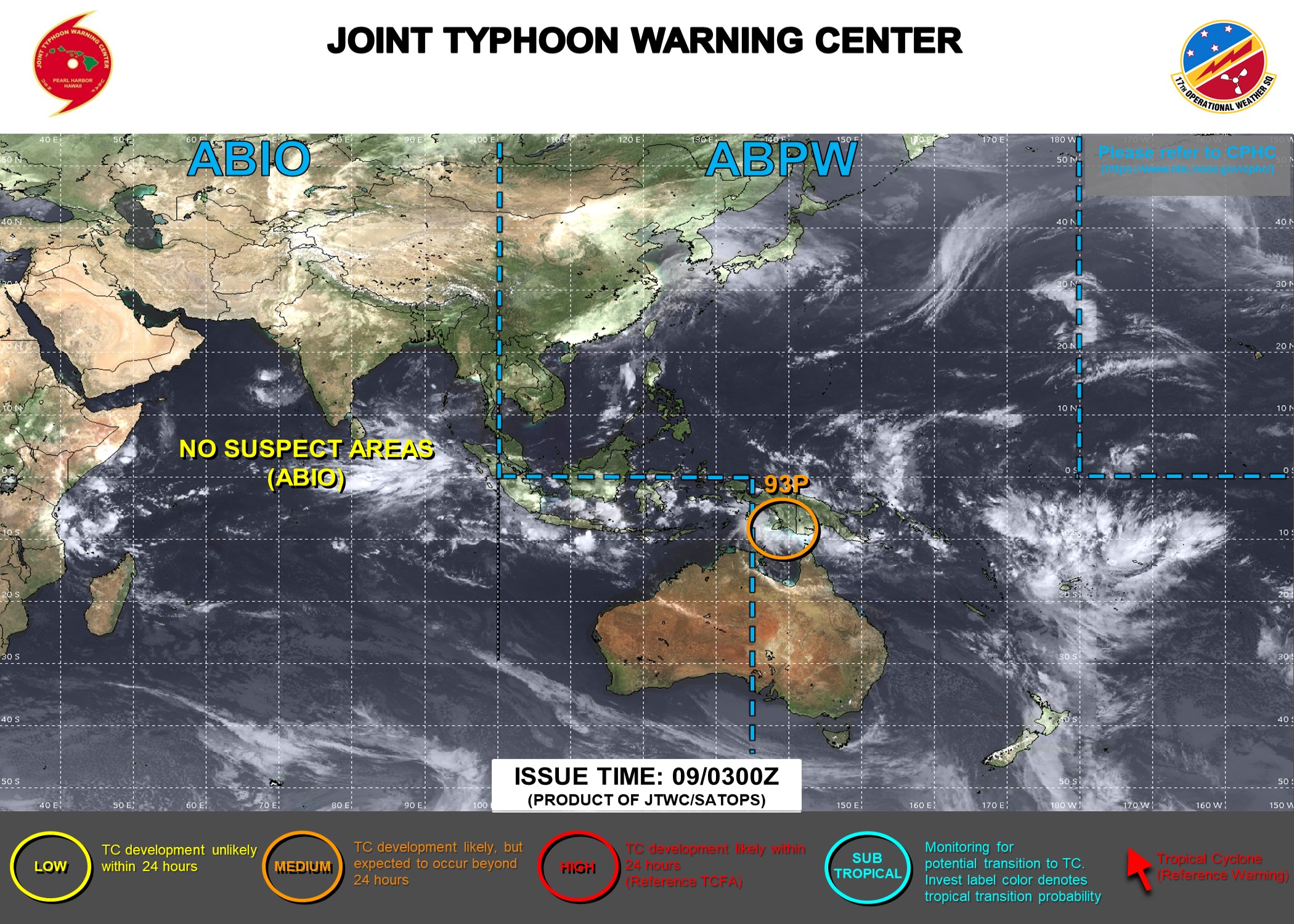
JTWC Tropical Warnings#
https://www.metoc.navy.mil/jtwc/jtwc.html
Forecast Parameters#
# project database
p_data = r'/media/administrador/HD2/SamoaTonga/data'
p_db = r'/media/administrador/HD1/DATABASES'
#site = 'Savaii' # Savaii / Upolu / Tongatapu
# (optional) get site from config file
nb_args = read_config_args(op.abspath(''), '09a_rainfall_tc_flooding')
site = nb_args['site']
print('Study site: {0}'.format(site))
Study site: Upolu
# noaa tmpfsc: automated forecast date, updated up to 3/4 days before present
years, months, days, runs = webscan_noaa_ncei()
y, m, d, h = int(years[-1]), int(months[-1]), int(days[-1]), int(runs[-1])
date_noaa = '{0:04d}{1:02d}{2:02d}'.format(y, m, d)
print('last available date for noaa tmpfsc: {0}'.format(date_noaa))
# get available run (NOAA prmsl forecast run [0, 6, 12, 18])
runs = [int(r) for r in runs]
# selected run to solve
run_sel = runs[0]
print('run: {0}'.format(run_sel))
# current date (for execution folder)
date = datetime.today().strftime('%Y%m%d%H%M')
print('forecast execution date: {0}'.format(date))
last available date for noaa tmpfsc: 20220909
run: 0
forecast execution date: 202209130650
# forecast parameters
_clean_forecast_folder_days = 7 # days to keep after forecast folder cleaning
# rainfall footprint
run_rainfall_footprint = True
# sst data download options
_load_local_cache = False
_clean = False
# choose agencies for TC warning
ags_sel = [
' UKMI', ' JMA', ' JGSI', ' AVNI', ' HWFI', ' NVGI',
' ECM2', ' JTWC', ' JNVI', ' NVG2', ' UEMI', ' UHM2', ' UKM2',
]
# Hazard winds
process_raster = False
# site related parameteres
if site == 'Savaii':
# site
site_ = site.lower()
nm = 'sp'
# area of interest
area_interest = [[177, -165+360] , [-21.5, -7]]
area_interest_grid = [[187.11, 187.89],[-13.89, -13.38]]
elif site == 'Upolu':
# site
site_ = site.lower()
nm = 'up'
# area of interest
area_interest = [[177, -165+360] , [-21.5, -7]]
area_interest_grid = [[187.848, 188.63],[-14.078, -13.756]]
elif site == 'Tongatapu':
# site
site_ = site
nm = 'tp'
# area of interest
area_interest = [[179, -169+360] , [-23.6, -18.1]]
area_interest_grid = [[184.59, 185.041],[-21.31, -21.0275]]
area_interest_plot = [x for sl in area_interest for x in sl]
Database#
p_site = op.join(p_data, site)
# deliverable folder
p_deliv = op.join(p_site, 'd09_rainfall_forecast')
# Lisflood database
p_lf_db = op.join(p_db, 'Lisflood_Rainfall')
# Basins path
p_basins = op.join(p_deliv, 'basins', '{0}_db_fixed.shp'.format(nm))
# Som fitting
p_som_fit = os.path.join(p_deliv, 'som_fit_30x30_4d.nc')
# Parameters from classification
p_params_fit = os.path.join(p_deliv, 'params_fit_30x30_4d_clust.nc')
# Dem
p_dem = os.path.join(p_deliv, site_ + '_5m_dem.nc')
# LHS cases
p_lhs_subset = os.path.join(p_deliv, 'LHS_mda_sel_lisflood_volume_rain.nc')
# ibtracs coef folder
p_ibtracs_coef = op.join(p_deliv, 'TC_forecast')
Forecast Output Folder#
p_forecast = op.join(p_site, 'forecast', '09_rainfall_tc_inundation')
# noaa download folder
p_fore_dl_sfc = op.join(p_site, 'download_noaa', '{0}'.format(date_noaa), 'tmpsfc')
# forecast folder
p_fore_date = op.join(p_forecast, date)
print('forecast date code: {0}'.format(date))
# prepare forecast subfolders for this date
p_fore_warn = op.join(p_fore_date, 'warnings')
for p in [p_fore_date, p_fore_warn, p_fore_dl_sfc]:
if not op.isdir(p):
os.makedirs(p)
# clean forecast folder
clean_forecast_folder(p_forecast, days=_clean_forecast_folder_days)
# clean noaa download extra folder
clean_forecast_folder(p_fore_dl_sfc, days=_clean_forecast_folder_days)
forecast date code: 202209130650
deleted folder: /media/administrador/HD2/SamoaTonga/data/Upolu/forecast/09_rainfall_tc_inundation/202209010240
# load basin data
bas_raw = gpd.read_file(p_basins)
basins = Obtain_Basins(bas_raw, site)
Download TC warnings#
# download TC warnings
files_ash, files_bsh = download_warning(p_fore_warn, code_basin='sh')
At: 2022-09-13T04:50:22
Total number of active storms: 3
Total number of active storms in the sh: 0
# stop forecast if no warnings
if len(files_bsh) == 0:
raise ValueError('No warnings in the SH. Forecast execution cancelled.')
sys.exit()
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
/tmp/ipykernel_29298/2713157806.py in <module>
1 # stop forecast if no warnings
2 if len(files_bsh) == 0:
----> 3 raise ValueError('No warnings in the SH. Forecast execution cancelled.')
4 sys.exit()
5
ValueError: No warnings in the SH. Forecast execution cancelled.
# choose storm code
code = 0
file_active = files_ash[code]
file_best = files_bsh[code]
print('active TC: {0}'.format(op.basename(file_active)))
print('best track: {0}'.format(op.basename(file_best)))
# show available warning times
name_warning, available_times_warning = get_info_TCs_warning(file_active)
print('name warning: {0}'.format(name_warning))
print('last available warning time: {0}'.format(available_times_warning[-1]))
# use last available time warning
time_warning = str(available_times_warning[-1])
print('time warning: {0}'.format(time_warning))
TC Warning Output#
tc_name = str(name_warning)
p_fore_tc = op.join(p_fore_date, tc_name + '_forecast')
fore_code = 'forecast_{0}_{1}'.format(tc_name, site)
p_ref_tc = op.join(p_fore_tc, '{0}.nc'.format(fore_code))
p_ref_tc_time = op.join(p_fore_tc, '{0}_time.nc'.format(fore_code))
p_ref_tc_area = op.join(p_fore_tc, '{0}_area.nc'.format(fore_code))
p_ref_tc_area_time = op.join(p_fore_tc, '{0}_area_time.nc'.format(fore_code))
p_out_tracks = op.join(p_fore_tc, 'OUT_TRACKS')
# warnings
p_fore_save_track = op.join(p_fore_date, 'Forecast_{0}_time_{1}.nc'.format(name_warning, time_warning[:-3]))
p_fore_save_track_sel = op.join(p_fore_date, 'Forecast_{0}_time_{1}_selected.nc'.format(name_warning, time_warning[:-3]))
for p in [p_fore_tc, p_out_tracks]:
if not op.isdir(p):
os.makedirs(p)
# Load TRACKS
TRACKS = Load_TCs_Warning(file_active, time = time_warning)
# save warning TRACKS
TRACKS.to_netcdf(p_fore_save_track)
TRACKS = xr.open_dataset(p_fore_save_track)
Area#
Figure Explanation
The figures below show TC warning tracks at the study site.
Need visual inspection on wether the tracks are inside the area of interest.
If there are no tracks in this area, don’t continue running the notebook.
Plot_Tracks_Maxwinds(TRACKS);
Plot_Tracks_Maxwinds(TRACKS, area_extent=area_interest_plot);
Plot_Tracks_Maxwinds_Density(TRACKS, area_extent=area_interest_plot);
Agencies selection#
print('Number of tracks: {0}'.format(len(TRACKS.track.values)))
print('Available Agencies:\n{0}\n------------'.format(TRACKS.Agency.values))
c, ia, ib = np.intersect1d(TRACKS.Agency.values, ags_sel, return_indices=True)
print('Total number of tracks: {0}'.format(len(ia)))
print('Agencies from list available : ' + str(np.array(ags_sel)[ib]))
TRACKS = TRACKS.isel(track=ia)
TRACKS['Index'] = (('track'), ia)
if TRACKS.track.size > 0:
Plot_Tracks_Maxwinds_Density(TRACKS, area_extent=area_interest_plot);
if TRACKS.track.size > 0:
Plot_Tracks_Maxwinds_zoom(TRACKS, area_interest_plot);
Rainfall#
Rainfall footprint#
Download needed SST data
# download SST data
print('download tmpsfc. run number {0}'.format(run))
sst = download_noaa(
'tmpsfc',
y, m, d, h,
p_f = p_fore_dl_sfc,
load_local_cache = _load_local_cache, clean=_clean,
)
print('')
f_save = 'tmpsfc' + '.'+str(y)+'.'+str(m)+'.'+str(hours[hi])+'.daily.grb2.nc'
sst.to_netcdf(op.join(path_fore_dl_sfc,f_save))
# Process tracks and add sst for rainfall model
forecast_tracks(TRACKS, sst, p_ibtracs_coef, p_fore_date, tc_name)
Load needed files
To run the model, we need to load the data corresponding to the SOM clustering (som_fit dataset) and the parameters associated to each of the 900 clusters (params_fit dataset).
# load SOM parameteres
som_fit = xr.open_dataset(p_som_fit)
params_fit = xr.open_dataset(p_params_fit)
# run the model
if run_rainfall_footprint:
# init output holders
DSS, DSS_A = [], []
DST, DST_A = [], []
TCKS = []
for a in range(len(TRACKS.track)):
# load track generated by forecast_tracks()
tck = xr.open_dataset(op.join(p_fore_date, tc_name + str(a) + '.nc'))
# reforecast track rain
tck, dss, dst, dss_area, dst_area = Reforecast_Track_Rain(
tck, area_interest, area_interest_grid, tc_name, som_fit, params_fit,
)
DSS.append(dss)
DST.append(dst)
DSS_A.append(dss_area)
DST_A.append(dst_area)
TCKS.append(tck)
# join output
DSS = xr.concat(DSS, dim='track'); DSS.to_netcdf(p_ref_tc)
DST = xr.concat(DST, dim='track'); DST.to_netcdf(p_ref_tc_time)
DSS_A = xr.concat(DSS_A, dim='track'); DSS_A.to_netcdf(p_ref_tc_area)
DST_A = xr.concat(DST_A, dim='track'); DST_A.to_netcdf(p_ref_tc_area_time)
else:
# load reforecast track rain
DSS = xr.open_dataset(p_ref_tc)
DST = xr.open_dataset(p_ref_tc_time)
DSS_A = xr.open_dataset(p_ref_tc_area)
DST_A = xr.open_dataset(p_ref_tc_area_time)
Figure Explanation
The figures below show accumulated precipitation (mm) for each track.
right panel shows a map with the basins.
# Plot each track accumulated precipitation
for t in range(len(DSS.track)):
fig = plt.figure(figsize=[25,7])
gs = gridspec.GridSpec(1, 2, wspace=0.25)
ax = fig.add_subplot(gs[0],projection = ccrs.PlateCarree(central_longitude=180))
tc_total_rain(TCKS[t], DSS.isel(track=t), fig=fig, ax=ax);
ax1=fig.add_subplot(gs[1])#,projection = ccrs.PlateCarree(central_longitude=180))
tc_total_rain_site(TCKS[t], DSS_A.isel(track=t), figsize=[25,12], basins=basins, fig=fig, ax=ax1);
SWATH#
Figure Explanation
Accumualted precipitation (mm) basin maps.
fig = plt.figure(figsize=[25,8])
gs = gridspec.GridSpec(1, 1, wspace=0.25)
ax1 = fig.add_subplot(gs[0]) #,projection = ccrs.PlateCarree(central_longitude=180))
tc_total_rain_site(tck, DSS_A.max(dim='track'), figsize=[25,12], basins=basins, fig=fig, ax=ax1);
Probabilistic assessment#
lims_dss = [150, 200, 250, 300]
for l in lims_dss:
DSS['rain_prob'+str(l)+'mm'] = (('lat', 'lon'), np.where(DSS.rain>l,1,0).sum(axis=(0))/(len(DSS.track.values)))
Figure Explanation
Rainfall higher than threshold probability maps.
fig = plt.figure(figsize=[25, 20])
gs = gridspec.GridSpec(2,2, wspace=0.3)
for a in range(len(lims_dss)):
ax = fig.add_subplot(gs[a], projection = ccrs.PlateCarree(central_longitude=180))
tc_total_rain(
TCKS[t], DSS.isel(track=t),
var='rain_prob'+str(lims_dss[a])+'mm',
fig=fig, ax=ax,
);
lims_dssa = [100, 150, 200, 250]
for l in lims_dssa:
DSS_A['rain_prob'+str(l)+'mm'] = (('lat', 'lon'), np.where(DSS_A.rain>l,1,0).sum(axis=(0))/(len(DSS_A.track.values)))
fig = plt.figure(figsize=[20, 12])
gs = gridspec.GridSpec(2,2, wspace=0.25)
for a in range(len(lims_dssa)):
ax1 = fig.add_subplot(gs[a])
tc_total_rain_site(
tck, DSS_A, font_s=12,mk_s=1, text=False,
var='rain_prob'+str(lims_dssa[a])+'mm', basins=basins,
fig=fig, ax=ax1,
);
Flooding#
Flooding footprint#
# lhs subset
n_cases = 50
df_v = xr.open_dataset(p_lhs_subset).isel(index=range(n_cases))
subset_norm = np.vstack([
(df_v['V'].values-np.nanmean(df_v['V'].values))/np.std(df_v['V'].values),
(df_v['I*'].values-np.nanmean(df_v['I*'].values))/np.std(df_v['I*'].values),
]).T
# dem
dem = xr.open_dataset(p_dem)
if 'z' in dem.data_vars:
dem = dem.rename({'z':'elevation'})
res_f = 2
dem = dem.isel(x=np.arange(0, len(dem.x.values),res_f), y = np.arange(0, len(dem.y.values),res_f))
dem['elevation'] = (('y', 'x'), np.where(dem.elevation.values < 0, np.nan, dem.elevation.values))
Figure Explanation
This figure show the basins maps.
# plot area basins
Plot_Basins(basins, figsize = [20, 15]);
Figure Explanation
TODO: FIGURE EXPLANATION.
# iterate each track
for tk in range(len(DSS.track)):
print('track : ' + str(tk))
dst_area = DST_A.isel(track=tk)
dss_area = DSS_A.isel(track=tk)
# Calculate mean rain per basin
x_utm, y_utm, INFO = tc_total_rain_site(
TCKS[tk], dss_area,
figsize=[25,10], rain=0,
figp=False,
basins=basins, points=1,
#vmax=970,
)
dss_area['X_UTM'] = (('lon'), x_utm)
dss_area['Y_UTM'] = (('lat'), y_utm)
# Hyetographs per basin
rain, _, _ = Plot_Rain_Hyetograph_Basins(
dst_area, dss_area,
basins, INFO, site,
deltat=1, thr_plot=1, thr=5,
fgsize1 = [20,6], fgsize2 = [20,15],
)
# Normalize rain to find analogues
tc_rain_norm = np.vstack([
(rain['Volume'].values-np.nanmean(df_v['V'].values))/np.std(df_v['V'].values),
(rain['I'].values-np.nanmean(df_v['I*'].values))/np.std(df_v['I*'].values),
]).T
# Analogue calculations
n_an = 3 # Number of analogue points
AN = Get_Analogues(n_an, tc_rain_norm, subset_norm, df_v)
# TODO
# Plot_Analogues(df_v, rain, AN, basins, figsize=[12,12])
OUT = Get_Flood_All_Basins(basins, AN, p_lf_db, nm)
# store track output
np.savetxt(op.join(p_out_tracks, 'Flooding_Metamodel_Analogues_{0}_{1}_xyz_points.nc'.format(tc_name, tk)), OUT.values)
OUT.dropna().set_index(['x','y']).to_xarray().to_netcdf(op.join(p_out_tracks, 'Flooding_Metamodel_Analogues_{0}_{1}_raster.nc'.format(tc_name, tk)))
extent = [dem.x.min()-1000, dem.x.max()+1000, dem.y.min()-1000, dem.y.max()+1000]
Plot_Rain_Flood_Points(basins, OUT, dem=dem, extent=extent)
if tk==0:
OUT_C = OUT.dropna().set_index(['x','y'])
else:
OUT_C = pd.concat([OUT_C, OUT.dropna().set_index(['x','y'])], axis=0, join='outer')
OUT_C = OUT_C.groupby(['x', 'y'])['z'].max().to_frame()
OUT_C = OUT_C.reset_index()
# store env output
OUT_C.to_xarray().rename({'index':'point'}).to_netcdf(op.join(p_out_tracks, 'Flooding_Metamodel_Analogues_envelope.nc'))
SWATH#
Figure Explanation
TODO: FIGURE EXPLANATION.
Plot_Rain_Flood_Points(basins, OUT_C, dem=dem, extent=extent);
print("execution end: {0}".format(datetime.today().strftime('%Y-%m-%d %H:%M:%S')))